FIGURE 1.
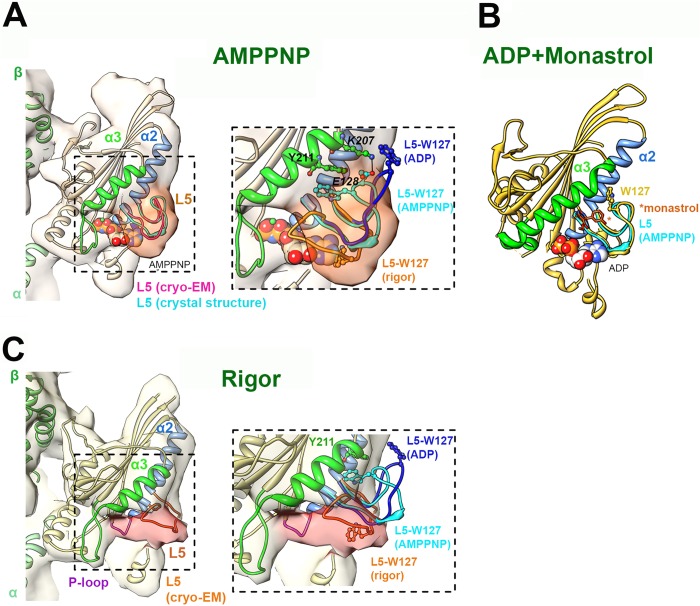
Nucleotide-dependent structural snapshots of the Eg5 loop L5. A, left, cryo-EM reconstruction (pale orange density; EMDB-2077) of the MT-bound human Eg5 motor domain bound to AMPPNP, viewed toward the nucleotide binding site (14). The density of L5 is highlighted in dark orange. The cryo-EM-derived docked coordinates (PDB entry 4AQV) are depicted in coral apart from the secondary structures adjacent to L5 (colored according to the labels). The loop L5 conformation of the Eg5·AMPPNP crystal structure (PDB entry 3HQD) (12) is superimposed in cyan for comparison. Right, enlarged view of loop L5. The conformation of L5 in the cryo-EM-derived rigor model (orange) and the ADP crystal structure (dark blue; PDB entry 1II6) (13) are also depicted, with the position of Trp-127 shown in a ball and stick representation in each. The residue Tyr-211 in the Eg5·AMPPNP crystal structure is also shown. B, monastrol/ADP-bound crystal structure of the human Eg5 motor domain (PDB entry 1Q0B) viewed toward the nucleotide binding site. Secondary structures adjacent to L5 are colored according to the labels, and monastrol is shown in orange sticks, indicated by an asterisk. Note the extended conformation of helix α3 (green) compared with its short conformation in the MT-bound motor. C, left, cryo-EM reconstruction (pale yellow density; EMDB-2078) of the MT-bound human Eg5 motor domain in the absence of nucleotide (rigor), viewed toward the nucleotide binding site. The density of L5 is highlighted in pink. The cryo-EM-derived docked coordinates (PDB entry 4AQW) are depicted in yellow apart from the secondary structures adjacent to L5 (colored according to the labels). Right, enlarged view of loop L5. The conformations of L5 in the AMPPNP (cyan) and the ADP (dark blue) crystal structures are also depicted, with the position of Trp-127 shown in a ball and stick representation in each. In each panel, the MT plus-end is toward the top, and bound nucleotide is depicted in a space-filling representation. EM reconstructions were contoured according to the expected molecular weight of the complex, and figures were prepared using Chimera (27).