Summary
Asparagine-linked glycosylation of proteins by the oligosaccharyltransferase (OST) occurs when acceptor sites or sequons (N-x≠P-T/S) on nascent polypeptides enter the lumen of the rough endoplasmic reticulum. Metazoan organisms assemble two isoforms of the OST that have different catalytic subunits (STT3A or STT3B) and partially non-overlapping cellular roles. Potential glycosylation sites move past the STT3A complex, which is associated with the translocation channel, at the protein synthesis elongation rate. Here, we investigated whether close spacing between acceptor sites in a nascent protein promotes site skipping by the STT3A complex. Biosynthetic analysis of four human glycoproteins revealed that closely spaced sites are efficiently glycosylated by an STT3B-independent process unless the sequons contain non-optimal sequence features, including extreme close spacing between sequons (e.g. NxTNxT) or the presence of paired NxS sequons (e.g. NxSANxS). Many, but not all, glycosylation sites that are skipped by the STT3A complex can be glycosylated by the STT3B complex. Analysis of a murine glycoprotein database revealed that closely spaced sequons are surprisingly common, and are enriched for paired NxT sites when the gap between sequons is less than three residues.
Key words: Asparagine-linked glycosylation, Oligosaccharyltransferase, Endoplasmic reticulum
Introduction
Asparagine-linked glycosylation is one of the most common protein modification reactions in eukaryotic cells, occurring on N-(x≠P)-T/S/C consensus sequons on newly synthesized proteins in the lumen of the rough endoplasmic reticulum (RER). Transfer of the preassembled oligosaccharide (GlcNAc2Man9Glc3) from a dolichol pyrophosphate carrier to sequons is mediated by the oligosaccharyltransferase (OST). Although N-glycosylation is often described as a post-translational protein modification reaction, most N-linked glycans are added to ribosome-bound nascent polypeptides as the protein is passing through the protein translocation channel into the lumen of the RER. A cotranslational mode of N-linked glycosylation ensures that addition of glycan occurs before protein folding, thereby relieving the restriction that sequons reside on surface-exposed loops or within disordered protein segments that can access the OST active site (Lizak et al., 2011).
The OST is a hetero-oligomeric integral membrane protein in most eukaryotes, including plants, fungi and metazoans (Kelleher and Gilmore, 2006). The catalytically active STT3 subunit of the OST is homologous to monomeric OSTs that are expressed in trypanosomes, Archaea and certain proteobacteria (Kelleher et al., 2003; Wacker et al., 2002; Yan and Lennarz, 2002). With the exception of Caenorhabditis species, genomes from all sequenced metazoan organisms encode two STT3 proteins (STT3A and STT3B) that are incorporated into distinct OST complexes (Kelleher et al., 2003; Shrimal et al., 2013b) along with a shared set of accessory subunits (ribophorin I, ribophorin II, OST48, DAD1 and OST4). The STT3A isoform of the OST complex is associated with the protein translocation channel and mediates cotranslational glycosylation of proteins (Ruiz-Canada et al., 2009; Shibatani et al., 2005). Surprisingly, siRNA-mediated depletion of STT3A in HeLa cells does not cause hypoglycosylation of most glycoproteins because the STT3B complex can mediate cotranslational, as well as post-translocational, glycosylation of sequons that are skipped by the STT3A complex. Glycoprotein substrates that are remarkably sensitive to STT3A depletion (e.g. prosaposin, progranulin and transferrin) are characterized by a very high density of disulfides in the folded structure, small independent folding domains (prosaposin and progranulin) and a relatively rapid rate of folding and exit from the RER (Ruiz-Canada et al., 2009; Shrimal et al., 2013a; Shrimal et al., 2013b). Sequons located in the C-terminal 50 residues of glycoproteins are skipped by STT3A and post-translocationally modified by STT3B (Shrimal et al., 2013b). Mutations in the human STT3A and STT3B genes cause two newly identified forms of congenital disorders of glycosylation (STT3A-CDG and STT3B-CDG) (Shrimal et al., 2013a). Fibroblasts from patients with STT3A-CDG and STT3B-CDG have defects in N-linked glycosylation that resemble HeLa cells that have been treated with STT3A- or STT3B-specific siRNAs (Shrimal et al., 2013a).
Depletion of non-catalytic OST subunits (ribophorin I, OST48 or DAD1) that are shared by both the STT3A and STT3B complexes generally cause a global defect in N-linked glycosylation because accessory subunit loss reduces the stability of both the STT3A and STT3B complexes (Roboti and High, 2012; Ruiz-Canada et al., 2009; Wilson and High, 2007). Likewise, fibroblasts from CDG patients with a mutation in the DDOST gene, which encodes OST48, have a general defect in N-linked glycosylation (Jones et al., 2012).
Closely spaced sequons might be prone to skipping by STT3A because of the bulky nature of the attached glycan, the kinetics of protein synthesis and the low fold molar excess of the oligosaccharide donor relative to the OST in cells. Potential glycosylation sites will pass by the translocation channel associated STT3A complex at the protein synthesis elongation rate, which is roughly 5–6 residues per second in mammalian cells (Hershey, 1991). Dolichol-linked oligosaccharide pools are low (∼1–2 nmol/g of tissue) (Gao and Lehrman, 2002; Kelleher et al., 2001) relative to the amount of the OST (∼0.5 nmol/g) (Guth et al., 2004; Kelleher et al., 1992). When tested using in vitro translation systems supplemented with microsomal membranes, glycosylation of adjacent NxT sites was unfavorable compared with sequons that are separated by one or more intervening residues (Karamyshev et al., 2005), indicating that close spacing can reduce glycosylation efficiency.
Nonetheless, efficient glycosylation of closely spaced sites has been documented for several glycoproteins and is known to be important for protein function and cell surface expression. Glycosylation of two closely spaced sequons in blood coagulation factor X is needed to prevent premature clearance of factor X from the bloodstream (Guéguen et al., 2010). Certain membrane transport proteins including the neuronal glycine transporter GlyT2 (Martínez-Maza et al., 2001), the GABA transporter GAT1 (Cai et al., 2005) and the bile salt export pump ABCB11 (Mochizuki et al., 2007) each contain a cluster of three or four closely spaced sequons. Interestingly, two of the potential glycosylation sites in these clusters need to be modified to achieve normal cell surface expression and activity of GlyT2, GAT1 and ABCB11. The CFTR protein has a pair of closely spaced sites, both of which need to be modified for maximal cell surface expression (Glozman et al., 2009).
Here, we have combined bioinformatic analysis and biochemical methods to analyze glycosylation of closely spaced sequons in mammalian glycoproteins. A surprisingly high percentage of sequons in murine glycoproteins are located within 20 residues of a neighboring glycosylation site. Close spacing between sites reduces the efficiency of N-glycosylation when one or more of the following conditions are met: (1) the sequons are adjacent (e.g. NxTNxT); (2) the closely spaced sequons contain serine residues as the hydroxyamino acid (e.g. NxSANxS); (3) the sequons contain non-optimal x residues; or (4) the sequons are in tandem arrays [e.g. (NxTZ)4–5]. siRNA-mediated depletion of STT3B revealed that closely spaced sequons in human glycoproteins are primarily glycosylated by the STT3A complex in an N-terminal to C-terminal scanning direction. The STT3B complex plays a secondary role in glycosylation of closely spaced sites by increasing site occupancy for non-optimal sites. Bioinformatic analysis of glycoprotein sequences and experimentally verified glycopeptides from seven model eukaryotes indicates that our conclusions concerning the factors that determine the glycosylation efficiency of closely spaced sites in HeLa cells are generally applicable to glycosylation of closely spaced sites in diverse eukaryotes.
Results
Closely spaced sequons are common in murine glycoproteins
If glycosylation consensus sites (N-x≠P-T/S) were randomly distributed within glycoprotein sequences, the sequon density and the distance between sites would be strongly dependent upon the percentage of asparagine, threonine and serine residues in the protein sequence. Glycosylated asparagine residues are exposed on the surface of folded glycoproteins, hence their location within a primary sequence could not be entirely random. The observed sequon density in glycoproteins exceeds the density predicted from the amino acid composition owing to positive selection for NxT sequons by eukaryotes that have the ER glycoprotein quality control pathway (Cui et al., 2009).
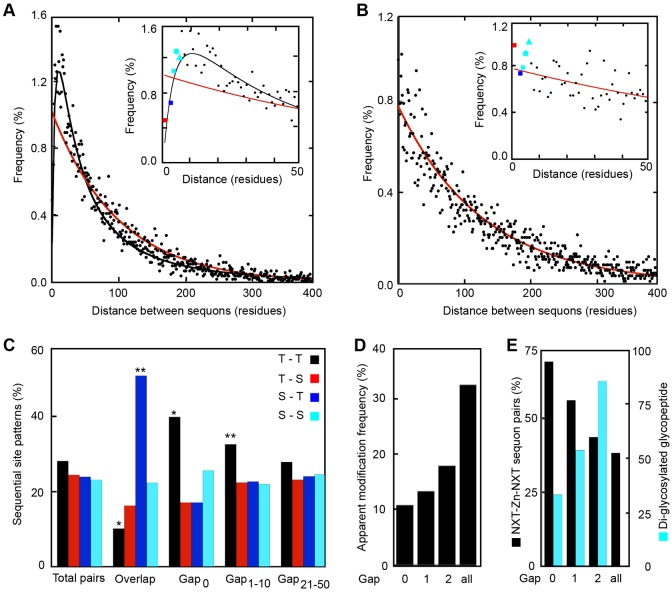
Using a database of experimentally verified glycopeptides (Zielinska et al., 2010) derived from 1902 murine glycoproteins we generated a collection of 11,983 N-x-T/S sequons (Shrimal et al., 2013b). Using this sequon collection, we determined the frequency distribution for distances between acceptor sites in murine glycoproteins (Fig. 1A). If glycosylation sites were uniformly distributed through glycoprotein sequences, the location of sites would be a Poisson process, with the distance between sites being described by an exponential density function. However, the observed distance between sequons fits a log normal distribution with a frequency maximum centered at ten residues between acceptor sites (Fig. 1A, black curve) rather than the distribution described by the exponential density function (Fig. 1A, red curve). Remarkably, more than 20% of all murine sequons are located less than 20 residues away from a neighboring sequon. Overlapping sites (NN-T/S-T/S; Fig. 1A, inset, red square) and adjacent sites (Nx-T/S-Nx-T/S; Fig. 1A, inset, blue square) are less abundant than expected for a uniform distribution (Fig. 1A, inset). However, sequon pairs with one- to three-residue gaps between the two sequons (e.g. Nx-T/S-Z1–3-Nx-T/S) are quite common (Fig. 1A inset, cyan square, circle and triangle, respectively).
Fig. 1.
Bioinformatic analysis of closely spaced sequons and glycans. (A,B) The distribution of distances in amino acid residues between asparagines in N-x≠P-T/S sites in murine glycoproteins (A) or in murine cytoplasmic proteins (B) was determined (black circles) and is plotted on a wide (0–400 residues) or narrow scale (0–50 residues, inset plot). The glycoprotein sequon database consisted of 11,983 N-x-T/S sequons in 1902 proteins. The murine cytoplasmic protein database had 10614 N-x≠P-T/S sites in 2256 proteins. Color-coded symbols in the inset plot correspond to overlapping sites (NN-T/S-T/S, red square), adjacent sites (e.g. NxT/S-NxT/S, blue square) or gap-1 to gap-3 sites (NxT/S-Z1–3-NxT/S; cyan square, circle and triangle, respectively). The frequency distribution for distances between sites was fit to a log normal distribution (A, black line) or an exponential density function (B, red line). (A) The expected distribution for uniformly distributed sites in the mouse glycoproteins is shown in red. (C) The observed distribution for overlapping sequons, adjacent sequons (Gap0) and sequon pairs with small (NxT/S-Z1–10-NxT/S) or intermediate gaps (NxT/S-Z21–50-NxT/S) was compared with the observed distribution of all sequon pairs. A pairwise chi-square test of association was performed to identify values that deviated from the total sequon pair distribution (*P<0.05; **P<0.005). (D) The apparent frequency of diglycosylated closely spaced sequons compared to the total apparent modification frequency. Apparent modification frequencies underestimate the actual modification frequency as a result of incomplete detection of all glycopeptides by mass spectrometry. (E) The number of diglycosylated glycopeptides (cyan bars) and the percentage of di-glycosylated glycopeptides (NxT/S-Z0–2-NxT/S) that have paired NxT sites (black bars) was determined for adjacent, gap-1 and gap-2 sites. The glycopeptide database consisted of 14091 glycopeptides identified by mass spectrometry from M. musculus, S. cerevisiae, S. pombe, A. thaliana, C. elegans, D. melanogaster and D. rerio.
A database of 2256 murine cytoplasmic proteins with a minimum of two N-x≠P-T/S sites per protein was analyzed in an identical manner (Fig. 1B). The observed distance between sites in cytoplasmic proteins was well fit by an exponential density function, indicating that NxT/S sequences in cytoplasmic proteins are uniformly distributed. As expected from the lower density of NxT sites in proteins that do not enter the secretory pathway (Cui et al., 2009), the mean distance between sites for the cytoplasmic proteins is greater (Fig. 1A,B, note change in the ordinate scale). Less than 15% of NxT/S sites were located within 20 residues of a neighboring site. Overlapping sites and adjacent sites in cytoplasmic proteins are not less abundant than expected for a uniform distribution (Fig. 1B, red and blue squares in the inset).
The percentage of sequon pairs that have the four possible combinations of threonine and serine residues was determined for overlapping sequons, adjacent sequons and sequons separated by small (NxT/S-Z1–10-NxT/S) or intermediate gaps (NxT/S-Z21–50-NxT/S). The observed composition of sequon pairs that are separated by 21–50 residues resembled the total sequon pair composition (Fig. 1C). The twofold enrichment of the NNST sequence among overlapping sequons primarily occurs at the expense of NNTT sequons. Overlapping sequons in glycoproteins are modified on a single asparagine residue (Karamyshev et al., 2005; Lockridge et al., 1987; Reddy et al., 1999; Reddy et al., 1988) owing to steric constraints within the OST active site (Lizak et al., 2011). The hydroxyamino acid (S or T) in the +2 position of an overlapping site influences which asparagine is glycosylated; NNST sequons favor modification of the second asparagine whereas NNTS sequons favor modification of the first asparagine (Reddy et al., 1999) because of the higher affinity of the OST for NxT sites than NxS sites (Bause, 1984). Of the 49 overlapping sequons in the murine sequon database, 16 have experimentally verified N-glycans (Zielinska et al., 2010), the majority of which are located on the second asparagine owing to the enrichment for overlapping NNST sequons. Overlapping sites in cytoplasmic proteins do not have a greater than expected proportion of NNST sequences (data not shown).
NxTNxT pairs were enriched among adjacent sequons relative to the expected distribution, whereas NxTNxS and NxSNxT showed modest decreases (Fig. 1C, Gap0). The enrichment for NxT sequon pairs decreases as the gap between sequons increases (Fig. 1C, Gap1–10 compared to Gap21–50). Cytoplasmic proteins showed no enrichment for NxT sequon pairs regardless of the gap between NxT sequences (data not shown). Glycopeptide databases from seven model organisms (Zielinska et al., 2010; Zielinska et al., 2012) were combined to examine the apparent modification frequency of closely spaced sequons relative to total sequons (Fig. 1D). Adjacent sequons, as well as gap-1 and gap-2 sequons, showed a low apparent diglycosylation frequency relative to the modification frequency for total sequons. Larger gap distances were not calculated because of the increasing probability that nearby sequons are located on separate tryptic or Glu-C glycopeptides. Diglycosylated adjacent and gap-1 sequons were strongly enriched in paired NxT sites (Fig. 1E). As the gap between sequons increased, the enrichment for paired NxT sites in diglycosylated peptides dropped to the predicted value for a random pattern of NxT and NxS sequons. Having established that closely spaced sequons in glycoproteins are common, and are enriched in NxT sites, we selected several human glycoproteins for biosynthetic analysis.
Efficient glycosylation of closely spaced NxT sequons
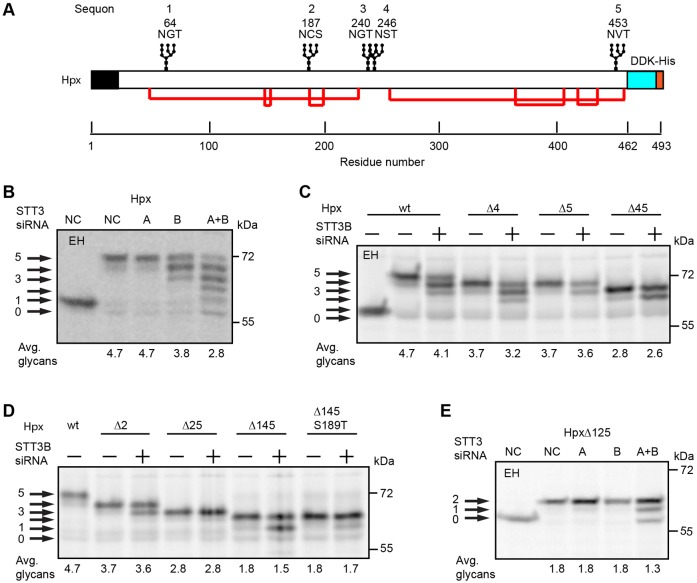
The secreted protein hemopexin (Hpx) has five glycosylation sites including a gap-3 sequon pair (N240GT and N246ST) and an extreme C-terminal sequon (Fig. 2A). The mobility difference between undigested Hpx and endoglycosidase-H-digested Hpx indicated the presence of multiple N-linked oligosaccharides (Fig. 2B). When the cellular content of STT3A and STT3B were both reduced by siRNA treatment, newly synthesized Hpx had 0–5 glycans. The most abundant Hpx glycoform synthesized by control cells was modified on all five sites. As observed previously (Ruiz-Canada et al., 2009), simultaneous depletion of STT3A and STT3B does not cause a complete block in N-glycosylation because total OST activity is only reduced by roughly fourfold when cells are treated with both siRNAs. Depletion of STT3B, but not STT3A, caused the synthesis of Hpx glycoforms that lacked one or two glycans indicating that Hpx contains two STT3B-dependent sites.
Fig. 2.
STT3B-independent glycosylation of closely spaced sites in hemopexin. (A) Diagram of hemopexin (Hpx) showing the signal sequence (black), glycosylation sites, disulfide bonds (red lines) and DDK-His tag. The five sequons are numbered 1–5; Hpx mutants lacking one or more sequons are designated as Hpx-ΔXYZ, where XYZ is the list of mutated sequon(s). HeLa cells were treated with negative control (NC) siRNA or siRNAs specific for STT3A or STT3B as indicated (B–E) for 48 hours prior to transfection with Hpx expression vectors. Cells were pulse labeled for 4 minutes and chased for 20 minutes. EH designates digestion with endoglycosidase H. Hpx glycoforms (0–5 glycans) were immunoprecipitated with anti-DDK sera and resolved by SDS-PAGE. Quantified values below gel lanes are for the displayed image that is representative of two or more experiments.
On the basis of our recent analysis of the role of STT3B in glycosylating extreme C-terminal sequons (Shrimal et al., 2013b), we predicted that sequon 5 in Hpx is an STT3B-dependent site. A series of Hpx glycosylation site mutants were analyzed to test this prediction (Fig. 2C) and to identify the second STT3B-dependent site (Fig. 2D). Glycosylation sites were eliminated by replacing the acceptor asparagine with glutamine (e.g. HpxΔ4 is HpxN246Q). Elimination of the N246ST site (HpxΔ4) reduced the number of attached glycans, but did not eliminate an STT3B-dependent site. By contrast, elimination of the C-terminal sequon either alone (HpxΔ5) or in combination with the fourth sequon (HpxΔ45) eliminated a single STT3B-dependent site (Fig. 2C). Mutagenesis of the second sequon (HpxΔ2) eliminated a second STT3B dependent site (Fig. 2D) indicating that the N187CS site is frequently skipped by STT3A. Analysis of two additional mutants (HpxΔ145 and HpxΔ145 S189T) revealed that site skipping of sequon 2 was reduced, but not eliminated, when the serine was replaced with threonine. Analysis of double (HpxΔ25) and triple (HpxΔ125) mutants that lack both STT3B sites showed that glycosylation of the two closely spaced sites (N240 and N246) was efficient and independent of STT3B, but could be mediated by STT3B when STT3A levels were reduced (Fig. 2D,E). Taken together, these experiments indicate that two NxT sites separated by a three-residue gap can be efficiently glycosylated by the translocation-channel-associated STT3A complex.
Skipping of adjacent NxS sites by the OST
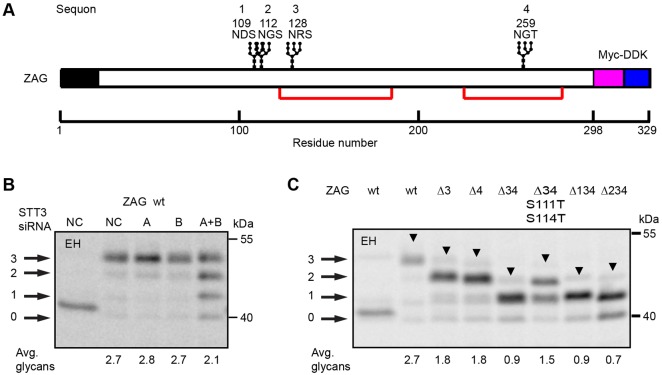
The secretory protein zinc α-2 glycoprotein (ZAG) has four glycosylation sites including two adjacent NxS sites (Fig. 3A, sequons 1 and 2). Although ZAG purified from human serum has three N-linked glycans (Araki et al., 1988; Sánchez et al., 1999), there is a disagreement concerning which site (N109 or N112) lacks an N-linked glycan. However, a glycopeptide containing modifications on both N109 and N112 has been identified by glycoproteomic analysis of ZAG obtained from human saliva (Ramachandran et al., 2006) and the MDA-MB-453 breast cancer cell line (Whelan et al., 2009). Wild-type ZAG and a panel of glycosylation site mutants were constructed to determine whether all four sites are glycosylated when ZAG is expressed in HeLa cells. Simultaneous depletion of STT3A and STT3B yielded four equally spaced products, one of which co-migrated with endoglycosidase-H-digested ZAG, indicating that ZAG synthesized by HeLa cells has three, not four glycans (Fig. 3B). Depletion of a single OST isoform had no obvious impact upon ZAG glycosylation. Mutagenesis of the third or fourth sequons (Δ3 or Δ4) caused an increase in ZAG mobility, showing that both sequons are modified in wild-type ZAG (Fig. 3C). The combination of the two mutations (ZAGΔ34) yielded a monoglycosylated protein unless the serine residues in the adjacent NxS sequons were replaced with threonine residues (ZAGΔ34 S111T S114T). However, glycan occupancy of one or both of the adjacent sites in the ZAGΔ34 S111T S114T mutant was clearly incomplete. The two ZAG triple mutants (ZAGΔ134 and ZAGΔ234) yielded monoglycosylated ZAG as the major product, indicating that both sequons could be glycosylated when tested alone. Even though the N109DS site showed lower glycan occupancy than the N112GS site when tested in isolation, we cannot conclude that the N109DS site is preferentially skipped in wild-type ZAG. In the context of ZAG, adjacent NxS sequons are poorly glycosylated even when both sites are functional.
Fig. 3.
Sequon skipping of adjacent NxS sites. (A) Diagram of ZAG showing the signal sequence (black), glycosylation sites, disulfide bonds (red lines) and Myc-DDK tag. The four sequons are numbered 1–4; ZAG mutants lacking one or more sequons are designated as ZAG-ΔXYZ, where XYZ is the list of mutated sequon(s). (B) HeLa cells were treated with negative control (NC) siRNA or siRNAs specific for STT3A or STT3B for 48 hours prior to transfection with wild-type ZAG. Cells expressing wild-type ZAG (B), or various ZAG glycosylation site mutants (C) were pulse labeled for 4 minutes and chased for 20 minutes. ZAG glycoforms (0–3 glycans) were immunoprecipitated using anti-DDK antibody and resolved by SDS-PAGE. Arrowheads designate a transient form of ZAG (see supplementary material Fig. S1). EH designates digestion with endoglycosidase H. Quantified values below gel lanes are for the displayed image that is representative of two or more experiments.
In addition to the major ZAG product detected in each lane, we detected an additional slower-migrating product (Fig. 3C, arrowheads) even when the protein had a single potential glycosylation site (e.g. ZAGΔ134). Pulse-labeling experiments and endoglycosidase H digestions show that the slow-mobility ZAG product is not explained by the presence of an additional N-linked glycan (supplementary material Fig. S1) because this product is prominent in pulse-labeled samples, but has a reduced intensity after the chase incubation. Slow cleavage of the ZAG signal sequence is one potential explanation for the transient slow mobility product.
Impact of hydroxyamino acid and gap length on sequon skipping
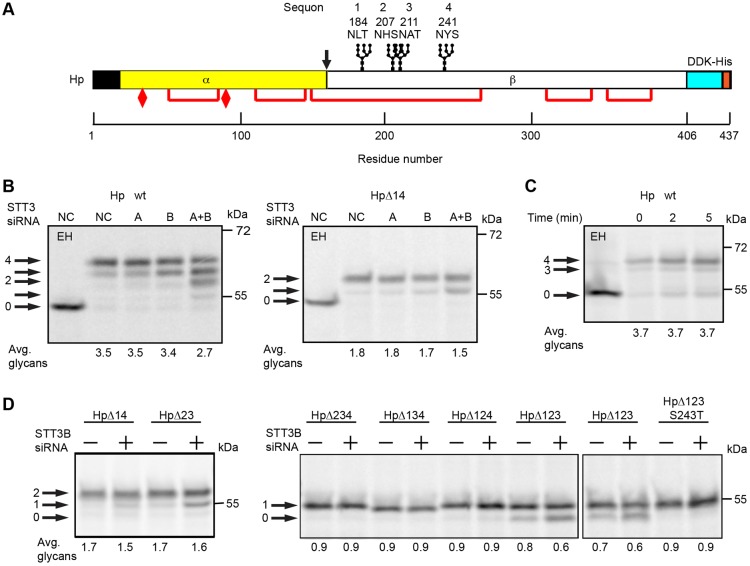
The secretory protein haptoglobin (Hp) is an α2β2 disulfide-linked tetramer that has four sequons in the β-subunit (Fig. 4A) including a gap-1 sequon pair (N207HS and N211AT). Although it is well established that haptoglobin is glycosylated on all four sites (Piva et al., 2002), the role of OST isoforms in haptoglobin glycosylation has not been investigated. Hp synthesized by control HeLa cells had on average 3.5 N-linked glycans (Fig. 4B). Depletion of STT3B, but not STT3A, reduced the percentage of fully glycosylated Hp slightly, suggesting that modification of one or more sequons is partially dependent upon STT3B. Pulse labeling of the HpΔ14 mutant indicated that glycosylation of the closely spaced sites was quite efficient and at most weakly dependent upon STT3B. A pulse-chase experiment (Fig. 4C) using wild-type Hp did not provide evidence for post-translational glycosylation of any sequon in Hp, unlike previously characterized STT3B-dependent sites in factor VII and sex hormone binding globulin (Ruiz-Canada et al., 2009; Shrimal et al., 2013b). Additional Hp glycosylation site mutants were analyzed in control and STT3B-depleted cells to identify the STT3B-dependent site. As observed for HpΔ14, glycosylation of HpΔ23 was slightly reduced in STT3B-depleted cells (Fig. 4D). Pulse labeling of the four possible triple mutants (e.g. HpΔ234) revealed that glycosylation of the fourth sequon (N241YS) was incomplete and partially STT3B dependent. The STT3B-dependence and incomplete modification of sequon 4 was eliminated by replacing serine 243 with a threonine residue (HpΔ123 S243T).
Fig. 4.
Glycosylation of haptoglobin. (A) Diagram of the haptoglobin precursor (Hp) showing the signal sequence (black), protease processing site at the α-β junction (arrow), glycosylation sites, disulfide bonds (red lines), cysteine residues that form interchain disulfides to link two α-subunits (elongated diamonds) and a DDK-His tag. The four sequons are numbered 1–4; Hp mutants lacking one or more sequons are designated as HpΔXYZ, where XYZ is the list of mutated sequon(s). (B,D) HeLa cells were treated with negative control (NC) siRNA or siRNAs specific for STT3A or STT3B as indicated for 48 hours prior to transfection with Hp expression constructs. Cells were then pulsed for 4 minutes and chased for 20 minutes. (C) Cells expressing Hp-wt were pulse labeled for 4 minutes and chased as indicated. Hp glycoforms were precipitated with anti-DDK sera and resolved by SDS-PAGE. EH designates digestion with endoglycosidase H. Quantified values below gel lanes are for the displayed image that is representative of two or more experiments.
The HpΔ14 mutant provided an excellent model protein to test the influence of the sequon type (NxT versus NxS), gap distance and OST isoform expression upon glycosylation of closely spaced sites. When the HpΔ14 derivative contained two adjacent NxT sequons (NHTNAT), both sites were efficiently glycosylated in control cells (Fig. 5A). Glycosylation of the HPΔ14 NHTNAT was reduced by 10% when STT3B was depleted indicating that one of the adjacent sites was infrequently skipped by STT3A. Glycosylation of the closely spaced NxT sites was independent of STT3B when a single alanine residue was inserted between the two sequons.
Fig. 5.
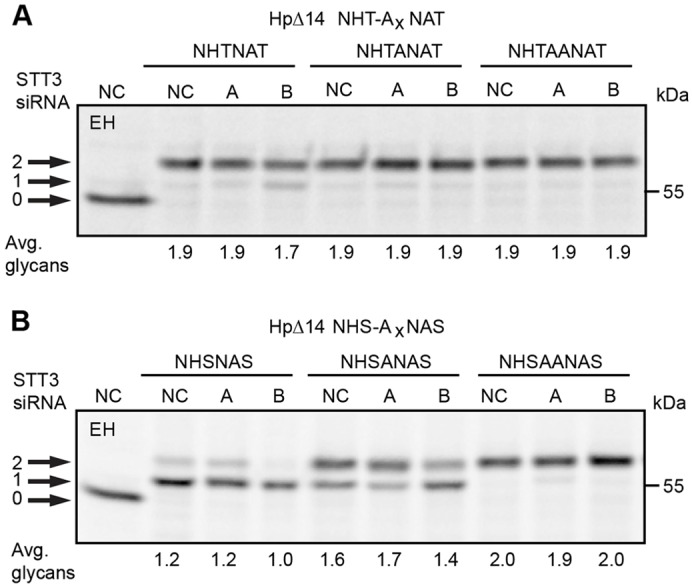
Sequence and gap-length dependence of sequon skipping. HeLa cells were treated with negative control (NC) siRNA or siRNAs specific for STT3A or STT3B as indicated for 48 hours prior to transfection with Hp expression constructs. The HpΔ14 derivatives had closely spaced NxT sites (A) or NxS sites (B) separated by 0–2 alanine residues. HeLa cells expressing Hp glycosylation site mutants were pulsed for 4 minutes and chased for 20 minutes. EH designates digestion with endoglycosidase H. Quantified values below gel lanes are for the displayed image that is representative of two or more experiments.
When the HpΔ14 NHSNAS construct was tested, Hp chains had an average of 1.2 glycans (Fig. 5B). Diglycosylated HpΔ14 NHSNAS was eliminated when STT3B levels were reduced. Insertion of a single alanine residue (HpΔ14 NHSANAS) enhanced modification of the NxS sites in control and STT3A-depleted cells. Addition of the second glycan remains partially dependent upon STT3B. Insertion of a second alanine residue results in STT3B-independent glycosylation of both NxS sites. Sequon skipping by the translocation-channel-associated STT3A complex occurs more frequently for NxS sequons and decreases quickly as the gap between sequons increases.
Sequon skipping within a tandem array
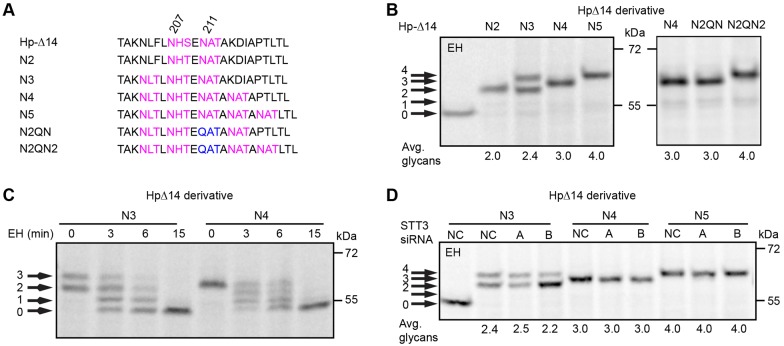
A series of HpΔ14 derivatives were constructed to determine whether there are additional factors that might limit glycosylation of closely spaced NxT sequons (Fig. 6A). The Hp-N2 construct, which has a gap-1 sequon pair, was glycosylated at both sites as expected (Fig. 6B). Addition of a third sequon (Hp-N3) resulted in the synthesis of two products, neither one of which co-migrated precisely with Hp-N2. Limited endoglycosidase H digestion of Hp-N3 indicates that the slower-migrating product has three N-linked oligosaccharides (Fig. 6C). Pulse-labeling of HpΔ14 derivatives that have four (Hp-N4) or five (Hp-N5) sequential NxT sites yielded single products displaying stepwise decreases in gel mobility relative to Hp-N2, consistent with the presence of three and four glycans, respectively (Fig. 6B). Limited Endo H digestion of Hp-N4 confirmed the presence of three N-linked glycans (Fig. 6C). Migration of Hp-N4 and Hp-N5 as a single product suggests that one sequon was uniformly skipped as opposed to incomplete modification of two or more sequons in the tandem array. Incomplete modification of two or more sites should yield a more complex pattern with multiple glycoforms. To test this hypothesis, two additional mutants were constructed (Hp-N2QN and N2QN2) wherein the third site was inactivated by the N211Q mutation. Consistent with the hypothesis that the third sequon in N4 and N5 is always skipped, the Hp-N4 product co-migrated with the Hp-N2QN product, but more rapidly than the Hp-N2QN2 product. How can we explain uniform skipping of the N211AT sequon in the Hp-N4 and Hp-N5 proteins, yet observe partial modification of all three sites in the Hp-N3? To address this question we expressed the Hp-N3, Hp-N4 and Hp-N5 constructs in HeLa cells that have reduced levels of STT3A or STT3B (Fig. 6D). Remarkably, glycosylation of the Hp-N4 and Hp-N5 proteins was completely insensitive to depletion of STT3B, indicating that the four modified sites in Hp-N5 are modified by STT3A, as Hp-N5 enters the ER lumen. By contrast, addition of the third glycan to the Hp-N3 protein is strongly reduced by depletion of STT3B, indicating that modification of third sequon in the array is unfavorable for both STT3A and STT3B. A reasonable interpretation of these results is that the steric bulk of neighboring N-linked glycans interferes with recognition of the N211AT site, hence this site is uniformly skipped in the Hp-N4 and Hp-N5 constructs.
Fig. 6.
Glycosylation of consecutive gap-1 sequons. (A) The sequences of HpΔ14 derivatives that have two to five closely spaced sites. (B–D) HeLa cells expressing Hp glycosylation site mutants were pulsed for 4 minutes and chased for 20 minutes. (C) Limited digestion of anti-DDK immunoprecipitates with endoglycosidase H (EH, 0–15 minutes). (D) HeLa cells were treated with negative control (NC) siRNA or siRNAs specific for STT3A or STT3B as indicated for 48 hours prior to transfection with Hp expression constructs. Quantified values below gel lanes are for the displayed image that is representative of two or more experiments.
Glycosylation of closely spaced C-terminal sequons
Several of the preceding experiments indicated that the STT3B isoform of the OST complex has a role in glycosylation of sub-optimal closely spaced sites. Sex hormone binding globulin (SHBG) has two extreme C-terminal glycosylation sites that are exclusively glycosylated by STT3B after the protein enters the ER lumen (Shrimal et al., 2013b). Using the previously characterized N380Q mutant as a model protein, we designed constructs to test whether STT3B could efficiently glycosylate closely spaced C-terminal sequons (Fig. 7A). As observed previously (Shrimal et al., 2013b), heterogeneous glycosylation of wild-type SHBG is primarily explained by incomplete modification of the N380RS site (Fig. 7B). Glycosylation of a gap-2 sequon pair in SHBG was very inefficient when the inserted sequon was NVS, but improved when the added sequon was NVT (Fig. 7B, NS-NT and NT-NT). Glycosylation of the SHBG NT-NT protein was less efficient than glycosylation of gap-2 NxT or NxS sequons in HpΔ14 (Fig. 5). Because N-glycosylation of sequons can be influenced by the x residue as well as flanking sequences, we tested whether the inserted NVT sequon was an inherently sub-optimal site. Glycosylation of the NVT sequon (Fig. 7B, NT-QT) was as efficient as glycosylation of the N396GT site. When tested separately, site occupancy for the N396GT site and the inserted NVT site were both 0.8 glycans/sequon. If glycosylation of these two C-terminal sites is independent, we would expect an average of 1.6 glycans for the SHBG NT-NT construct. Instead, the observed value of 1.3 glycans per SHBG NT-NT indicates that modification of one C-terminal site reduces subsequent glycosylation of the second site.
Fig. 7.
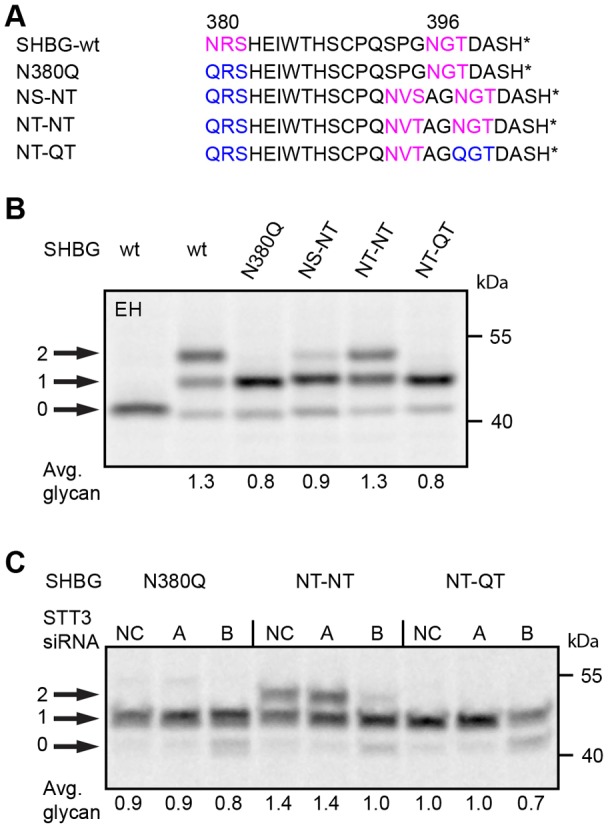
Glycosylation of extreme C-terminal closely spaced sites. (A) The C-terminal segment of sex hormone binding globulin (SHBG) and glycosylation site mutants. The asterisk designates the C-terminus. HeLa cells that were untreated (B) or treated with negative control (NC) siRNA or siRNAs specific for STT3A or STT3B for 48 hours (C) were transfected with SHBG expression vectors and radiolabeled 24 hours later using a 4 minute pulse label followed by a 20 minute chase. SHBG glycoforms (0–2 glycans) were immunoprecipitated with anti-SHBG and resolved by SDS-PAGE. EH designates digestion with endoglycosidase H. Quantified values below gel lanes are for the displayed image that is representative of two or more experiments.
Glycosylation of both C-terminal sites in wild-type SHBG is an STT3B-dependent process (Shrimal et al., 2013b). HeLa cells that had been treated with siRNAs specific for STT3A or STT3B were transfected with the SHBG N380Q, NT-NT or NT-QT constructs to verify that the inserted site NVT site was also STT3B dependent (Fig. 7C). Depletion of STT3A had no effect upon glycosylation of any of the SHBG derivatives. Depletion of STT3B reduced glycosylation of the N380Q mutant and the NT-QT mutant, indicating that both sites are glycosylated by STT3B. Glycosylation of the SHBG NT-NT protein was also reduced in STT3B-depleted cells; hence, we can conclude that closely spaced C-terminal sites are not efficiently modified by STT3B.
Discussion
Sequon skipping by the translocation-channel-associated STT3A complex
The Campylobacter lari PglB:acceptor peptide co-crystal structure revealed how sequons are recognized and positioned within the OST active site (Lizak et al., 2011). A remarkable feature of the OST active site is that the peptide-binding motifs that recognize the sequon reside on one face of PglB, whereas the donor substrate binding site and catalytic residues are exposed on the opposite face with the asparagine side chain of the acceptor peptide projecting through a narrow porthole that connects the two surfaces. Glycopeptide exit from the catalytic site must occur after the porthole is opened by the movement of a large flexible loop linking two of the transmembrane spans of PglB. The glycosylated sequon has to detach from the peptide-binding motif to permit the OST to scan the nascent polypeptide for the next acceptor site. Pulse labeling of hemopexin and derivatives thereof revealed very efficient STT3B-independent glycosylation of paired NxT sites that are separated by a three residue gap (e.g. NGT-GHG-NST). Paired NxT sites were also diglycosylated by STT3A in haptoglobin when separated by a one or two residue gap. These results indicate that a glycosylated asparagine can exit the active site, the glycan can be flipped to face away from the peptide binding face of STT3A and a nearby NxT site can be successfully positioned within the peptide-binding cleft. The rapid elongation of the polypeptide and the low abundance of the donor substrate do not pose a challenge to efficient cotranslational modification of closely spaced NxT sites by STT3A.
Local sequence context effects that are likely to reduce glycosylation of adjacent or gap-1 NxT sites include sub-optimal x residues (Kasturi et al., 1995) as well as proline residues located at the +3 position relative to an asparagine acceptor site (Gavel and von Heijne, 1990). The incomplete modification of the ZAGΔ34 S111T S114T mutant is probably explained by the negative impact of the sub-optimal x residue in the NDT sequon. The remarkable enrichment of paired NxT sites among adjacent sequons provides support for positive selection of optimal sequons in adjacent sites. The enrichment of diglycosylated NxTNxT and NxT-Z1-NxT peptides indicates that our observations concerning the modification efficiency of closely spaced sites in haptoglobin-derived substrates are applicable to modification of closely spaced sites in glycoproteins from diverse eukaryotic organisms.
Enzyme kinetics studies have shown that the eukaryotic and eubacterial OSTs have a higher affinity for synthetic peptides with NxT sequons than NxS sequons (Bause, 1984; Gerber et al., 2013). For that reason, we anticipated that closely spaced NxS sequon pairs might be hypoglycosylated more frequently than NxT sequon pairs. Pulse labeling of ZAG, which has adjacent NxS sites, showed that only one of the two adjacent NxS sites was modified in HeLa cells. When each site was tested in isolation the N-terminal N109DS site (ZAGΔ234) was incompletely modified unlike the N112GS site. Glycosylation assays using canine pancreas membranes have shown that the certain amino acids (W, D, E and L) at the x position of NxS sequons strongly reduce glycosylation efficiency (Shakin-Eshleman et al., 1996). It is unclear why HeLa cell ZAG and human serum ZAG lack one of the glycans on the adjacent NxS sites whereas ZAG synthesized by two other cell types is modified on both the N109DS and N112GS sequons (Ramachandran et al., 2006; Whelan et al., 2009). It should be noted that glycoproteomic detection of the diglycosylated ZAG glycopeptide does not indicate that the predominant ZAG glycoform is modified on both sites. Differences in the cellular content of the STT3A or STT3B complexes or the dolichol-oligosaccharide donor pool might contribute to cell-type-specific differences in glycosylation efficiency. The adjacent and gap-1 NxS sites that were tested in the haptoglobin-derived model substrates also showed incomplete modification of closely spaced NxS sites.
Inefficient modification of closely spaced sites by STT3B
Previous studies indicate that STT3B-containing OST complexes are able to glycosylate certain sequons that are skipped by STT3A, either in a cotranslational mode or a post-translocational mode (Ruiz-Canada et al., 2009; Shrimal et al., 2013b). Here, we observed that STT3B contributes to the glycosylation of closely spaced sequons that are skipped by STT3A in haptoglobin derivatives with adjacent NxT sites or when the sequons contained serine residues at the +2 position (NHSNAS and NHSANAS). With the exception of the NHTNAT site, STT3B was only able to modify a subfraction of the skipped sites. It is not known how STT3B locates substrates that have skipped acceptor sites and modifies them in either a cotranslational or a post-translational mode. If substrate recognition by STT3B is diffusion driven, the incomplete modification of skipped sites by STT3B might indicate that the glycoprotein begins to fold before it encounters STT3B. Further evidence that the STT3B complex is not proficient at modification of closely spaced sites was obtained by analysis of SHBG derivatives, where we observed that glycosylation of an extreme C-terminal site interfered with subsequent modification of the nearby gap-2 site.
The analysis of tandem arrays of gap-1 NxT sites provided support for a scanning mechanism of acceptor site recognition. The third site within arrays that contained four or five NxT sites was uniformly skipped, consistent with a steric clash caused by the glycans attached to the −4 and −8 positions. Modification of the third site in the N3 array was incomplete and strongly dependent upon STT3B. Once the N211AT site in the N5 array was skipped, the fourth and fifth sites were efficiently modified in an STT3B-independent manner. Although the Hp tandem array constructs are artificial, our search of the HIV sequence database (http://www.hiv.lanl.gov/components/sequence/HIV/search/search.html) identified multiple gp120 sequences that have similar clusters of gap-1 sequons within variable loop 1.
STT3B-dependent sites in hemopexin and haptoglobin
Hemopexin contains two glycosylation sites that are STT3B dependent, hence frequently skipped by STT3A. As expected, the extreme C-terminal N453VT site is STT3B dependent. Multiple factors are likely to contribute to STT3A skipping of the N187CS sequon in Hpx because we observed that the STT3B dependence was reduced, but not eliminated by acceptor site optimization (Hpx S189T mutation). Complete modification of the N241YS site in haptoglobin was also dependent upon STT3B. In contrast to the N187CS site in hemopexin, replacing the serine residue with a threonine residue (HpΔ123 S243T mutant) reduced site skipping by STT3A to a level that was undetectable in STT3B-depleted cells. A reduction in STT3B dependence upon site optimization indicates that sequon skipping by STT3A is at least partly explained by the reduced affinity of NxS relative to NxT sequons. The internal STT3B-dependent sites in Hpx and Hp join a growing list of sequons that are skipped at high frequency by STT3A and are subsequently modified by STT3B.
The STT3B complex reduces glycoprotein heterogeneity
The presence of a NxT/S site within a protein coding sequence does not insure N-glycosylation even for proteins that enter the secretory pathway. Certain sequons are always skipped whereas other sequons show low glycan occupancy. Our analysis of closely spaced sites in human glycoproteins indicates that adjacent and gap-1 sites contribute to the phenomena of low glycan occupancy and uniformly skipped sites. The duplication of the OST catalytic subunit to obtain the STT3A and STT3B complexes allows cotranslational scanning of nascent polypeptides for acceptor sites by STT3A followed by cotranslational as well as post-translocational modification of skipped sites by STT3B. The observation that depletion of STT3A does not cause a global reduction in N-linked oligosaccharides indicates that the STT3B complex can modify most of the glycosylation sites we have tested unless the protein rapidly acquires a conformation that is not compatible with the enzyme active site. By serving as a failsafe for protein N-glycosylation, the STT3B complex reduces glycoprotein heterogeneity in terms of variable site occupancy.
Materials and Methods
Cell culture and plasmid or siRNA transfection
HeLa cells (ATCC CCL-13) were cultured in 10 cm2 dishes at 37°C in DMEM (GIBCO, Grand Island, NY), 10% fetal bovine serum with penicillin (100 units/ml) and streptomycin (100 µg/ml). HeLa cells were seeded at 30% confluency for siRNA transfection or 80% confluency for plasmid transfection in 60 mm dishes and grown for 24 hours prior to transfection with siRNA (60 nM NC; 50 nM STT3A; 60 nM STT3B) or plasmid (8 µg) and Lipofectamine 2000 in Opti-MEM (GIBCO) using a protocol from the manufacturer (Invitrogen, Grand Island, NY). Plasmid transfection was done after 48 hours of siRNA transfection and cells were assayed 24 hours later. The siRNAs specific for STT3A and STT3B were characterized previously (Ruiz-Canada et al., 2009). The STT3A siRNAs are 5′-GGCCGUUUCUCUCACCGGCdTdT-3′ annealed with 5′-UCCGGUGAGAGAAACGGCCdTdT-3′. The STT3B siRNAs are 5′-GCUCUAUAUGCAAUCAGUAdTdT-3′annealed with 5′-CACUGAUUGCAUAUAGAGCdTdT-3′. Negative control siRNA was purchased from Qiagen.
The Myc-DDK tagged ZAG expression vector was purchased from OriGene (Rockville, MD). Hemopexin was amplified from a cDNA clone (Origene), and cloned into pCMV6-AC-DDK-His vector (OriGene). The SHBG and N380Q SHBG mutants in the vector pRC/CMV (Invitrogen) were gifts from Dr Geoffrey Hammond (Child and Family Research Institute; Vancouver British Columbia, Canada). A haptoglobin cDNA clone was obtained from Dr Kylie Walters (University of Minnesota) and cloned into pCMV6-AC-DDK-His vector (OriGene). Site-directed mutagenesis was used to insert or eliminate glycosylation sites. All glycosylation sites were inactivated by asparagine to glutamine substitutions.
Radiolabeling and immunoprecipitation of glycoproteins
Cell culture medium was replaced with methionine and cysteine-free DMEM medium (GIBCO), containing 10% dialyzed fetal bovine serum 20 minutes prior to the addition of 200 µCi/ml of Tran35S label (Perkin Elmer, Waltham, MA). Pulse-labeling periods were terminated by the addition of unlabeled methionine (3.75 mM) and cysteine (0.75 mM). Cells from one culture dish at each time point were lysed at 4°C by a 30 minute incubation with 1 ml of RIPA lysis buffer and 1× PIC [protease inhibitor cocktail, as defined previously (Kelleher et al., 1992)]. Cell lysates were clarified by centrifugation (2 minutes at 13,000 rpm), and precleared by incubation for 2 hours with control IgG and a mixture of Protein-A/G Sepharose beads (Zymed Laboratories, San Francisco, CA). The precleared lysates were incubated overnight with protein- or epitope-tag-specific antibodies followed by the addition of a second aliquot of Protein-A/G Sepharose beads and incubated for 4 hours. Beads were washed five times with RIPA lysis buffer and twice with 10 mM Tris-HCl before eluting proteins with gel loading buffer. Antibodies were obtained from the following sources: anti-DDK (anti-FLAG; Sigma, St. Louis, MO) and anti-SHBG (R&D Systems, Minneapolis, MN). As indicated, immunoprecipitated proteins were digested with endoglycosidase H (New England Biolabs, Ipswich, MA). Dry gels were exposed to a phosphor screen (Fujifilm, Tokyo, Japan) and scanned in Typhoon FLA 9000 (G.E. Healthcare, Pittsburgh, PA, USA) and quantified using AlphaEase FC (Santa Clara, CA).
Bioinformatics analysis of the murine glycoproteomic database
A murine glycopeptide database (4922 glycopeptides in 1902 glycoproteins) was derived from the high confidence (Class I) murine N-glycosylation sites identified by mass spectrometry (Zielinska et al., 2010) after exclusion of 130 glycopeptides as described (Shrimal et al., 2013b). The glycoprotein sequences were downloaded, and the locations of the glycopeptides were verified. A complete list of 11,983 sequons [N-(x≠P)-T/S] in the 1902 murine proteins was generated. NxC sequons were excluded because of their much lower modification frequency than NxT or NxS sequons.
The distance between sequons in the murine glycoproteins was determined after excluding 129 proteins with a single sequon. Each pair of sequons for which a distance was calculated was further categorized as a NxT-NxT, NxT-NxS, NxS-NxT or NxS-NxS pair depending upon the sequence of the two sites.
The frequency distribution for distance between sequons was fit to the following equation: 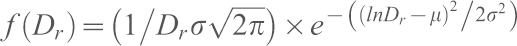 , where Dr is the distance between sequons in residues, μ is the mean and σ the s.d. of the natural logarithm of Dr.
, where Dr is the distance between sequons in residues, μ is the mean and σ the s.d. of the natural logarithm of Dr.
The curve for a random distribution of sequons in murine glycoproteins was obtained by using a random number generator to reassign the location of sequons in each of the murine glycoproteins. The distance between randomly assigned sequons was fit to the exponential density function:
where μ is the mean distance between randomly assigned sequons.
The murine glycopeptide database was combined with class I glycopeptides from six other model organisms (S. cerevisiae, S. pombe, A. thaliana, C. elegans, D. melanogaster and D. rerio) (Zielinska et al., 2012) to obtain a database with 14091 experimentally verified glycopeptides. The combined glycopeptide database was used to generate Fig. 1D,E.
A database of murine cytoplasmic proteins was obtained by searching the UniProt database for mouse cytoplasmic proteins. A total of 3915 unique protein sequences were downloaded for analysis. Of the 3915 proteins, 842 lacked any N-x≠P-T/S sequences, whereas 817 had a single N-x≠P-T/S sequence. After elimination of these 1659 proteins, we had a collection of 2256 cytoplasmic proteins that contained 10614 N-x≠P-T/S sequences. The distance between sites was calculated as described for the murine glycoproteins and fit to the exponential density function.
Supplementary Material
Acknowledgments
The authors thank Dr Geoffrey Hammond (Child and Family Research Institute; Vancouver British Columbia, Canada) and Dr Kylie Walters (University of Minnesota) for providing the SHBG and haptoglobin expression vectors, respectively.
Footnotes
Author contributions
S.S. and R.G. conceived, designed and interpreted experiments. S.S. performed all experiments. R.G. was responsible for bioinformatic analysis. S.S. and R.G. wrote the paper.
Funding
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health [grant number GM43768]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.139584/-/DC1
References
- Araki T., Gejyo F., Takagaki K., Haupt H., Schwick H. G., Bürgi W., Marti T., Schaller J., Rickli E., Brossmer R. et al. (1988). Complete amino acid sequence of human plasma Zn-alpha 2-glycoprotein and its homology to histocompatibility antigens. Proc. Natl. Acad. Sci. USA 85, 679–683 10.1073/pnas.85.3.679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bause E. (1984). Model studies on N-glycosylation of proteins. Biochem. Soc. Trans. 12, 514–517 [DOI] [PubMed] [Google Scholar]
- Cai G., Salonikidis P. S., Fei J., Schwarz W., Schülein R., Reutter W., Fan H. (2005). The role of N-glycosylation in the stability, trafficking and GABA-uptake of GABA-transporter 1. Terminal N-glycans facilitate efficient GABA-uptake activity of the GABA transporter. FEBS J. 272, 1625–1638 10.1111/j.1742-4658.2005.04595.x [DOI] [PubMed] [Google Scholar]
- Cui J., Smith T., Robbins P. W., Samuelson J. (2009). Darwinian selection for sites of Asn-linked glycosylation in phylogenetically disparate eukaryotes and viruses. Proc. Natl. Acad. Sci. USA 106, 13421–13426 10.1073/pnas.0905818106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N., Lehrman M. A. (2002). Analyses of dolichol pyrophosphate-linked oligosaccharides in cell cultures and tissues by fluorophore-assisted carbohydrate electrophoresis. Glycobiology 12, 353–360 10.1093/glycob/12.5.353 [DOI] [PubMed] [Google Scholar]
- Gavel Y., von Heijne G. (1990). Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 3, 433–442 10.1093/protein/3.5.433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber S., Lizak C., Michaud G., Bucher M., Darbre T., Aebi M., Reymond J. L., Locher K. P. (2013). Mechanism of bacterial oligosaccharyltransferase: in vitro quantification of sequon binding and catalysis. J. Biol. Chem. 288, 8849–8861 10.1074/jbc.M112.445940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozman R., Okiyoneda T., Mulvihill C. M., Rini J. M., Barriere H., Lukacs G. L. (2009). N-glycans are direct determinants of CFTR folding and stability in secretory and endocytic membrane traffic. J. Cell Biol. 184, 847–862 10.1083/jcb.200808124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guéguen P., Cherel G., Badirou I., Denis C. V., Christophe O. D. (2010). Two residues in the activation peptide domain contribute to the half-life of factor X in vivo. J. Thromb. Haemost. 8, 1651–1653 10.1111/j.1538-7836.2010.03905.x [DOI] [PubMed] [Google Scholar]
- Guth S., Völzing C., Müller A., Jung M., Zimmermann R. (2004). Protein transport into canine pancreatic microsomes: a quantitative approach. Eur. J. Biochem. 271, 3200–3207 10.1111/j.1432-1033.2004.04252.x [DOI] [PubMed] [Google Scholar]
- Hershey J. W. B. (1991). Translational control in mammalian cells. Annu. Rev. Biochem. 60, 717–755 10.1146/annurev.bi.60.070191.003441 [DOI] [PubMed] [Google Scholar]
- Jones M. A., Ng B. G., Bhide S., Chin E., Rhodenizer D., He P., Losfeld M. E., He M., Raymond K., Berry G. et al. (2012). DDOST mutations identified by whole-exome sequencing are implicated in congenital disorders of glycosylation. Am. J. Hum. Genet. 90, 363–368 10.1016/j.ajhg.2011.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamyshev A. L., Kelleher D. J., Gilmore R., Johnson A. E., von Heijne G., Nilsson I. (2005). Mapping the interaction of the STT3 subunit of the oligosaccharyl transferase complex with nascent polypeptide chains. J. Biol. Chem. 280, 40489–40493 10.1074/jbc.M509168200 [DOI] [PubMed] [Google Scholar]
- Kasturi L., Eshleman J. R., Wunner W. H., Shakin-Eshleman S. H. (1995). The hydroxy amino acid in an Asn-X-Ser/Thr sequon can influence N-linked core glycosylation efficiency and the level of expression of a cell surface glycoprotein. J. Biol. Chem. 270, 14756–14761 10.1074/jbc.270.24.14756 [DOI] [PubMed] [Google Scholar]
- Kelleher D. J., Gilmore R. (2006). An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R 10.1093/glycob/cwj066 [DOI] [PubMed] [Google Scholar]
- Kelleher D. J., Kreibich G., Gilmore R. (1992). Oligosaccharyltransferase activity is associated with a protein complex composed of ribophorins I and II and a 48 kd protein. Cell 69, 55–65 10.1016/0092-8674(92)90118-V [DOI] [PubMed] [Google Scholar]
- Kelleher D. J., Karaoglu D., Gilmore R. (2001). Large-scale isolation of dolichol-linked oligosaccharides with homogeneous oligosaccharide structures: determination of steady-state dolichol-linked oligosaccharide compositions. Glycobiology 11, 321–333 10.1093/glycob/11.4.321 [DOI] [PubMed] [Google Scholar]
- Kelleher D. J., Karaoglu D., Mandon E. C., Gilmore R. (2003). Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Mol. Cell 12, 101–111 10.1016/S1097-2765(03)00243-0 [DOI] [PubMed] [Google Scholar]
- Lizak C., Gerber S., Numao S., Aebi M., Locher K. P. (2011). X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355 10.1038/nature10151 [DOI] [PubMed] [Google Scholar]
- Lockridge O., Bartels C. F., Vaughan T. A., Wong C. K., Norton S. E., Johnson L. L. (1987). Complete amino acid sequence of human serum cholinesterase. J. Biol. Chem. 262, 549–557 [PubMed] [Google Scholar]
- Martínez-Maza R., Poyatos I., López-Corcuera B., N úñez E., Giménez C., Zafra F., Aragón C. (2001). The role of N-glycosylation in transport to the plasma membrane and sorting of the neuronal glycine transporter GLYT2. J. Biol. Chem. 276, 2168–2173 10.1074/jbc.M006774200 [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Kagawa T., Numari A., Harris M. J., Itoh J., Watanabe N., Mine T., Arias I. M. (2007). Two N-linked glycans are required to maintain the transport activity of the bile salt export pump (ABCB11) in MDCK II cells. Am. J. Physiol. 292, G818–G828 10.1152/ajpgi.00415.2006 [DOI] [PubMed] [Google Scholar]
- Piva M., Moreno J. I., Sharpe-Timms K. L. (2002). Glycosylation and over-expression of endometriosis-associated peritoneal haptoglobin. Glycoconj. J. 19, 33–41 10.1023/A:1022580813870 [DOI] [PubMed] [Google Scholar]
- Ramachandran P., Boontheung P., Xie Y., Sondej M., Wong D. T., Loo J. A. (2006). Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 5, 1493–1503 10.1021/pr050492k [DOI] [PubMed] [Google Scholar]
- Reddy V. A., Johnson R. S., Biemann K., Williams R. S., Ziegler F. D., Trimble R. B., Maley F. (1988). Characterization of the glycosylation sites in yeast external invertase. I. N-linked oligosaccharide content of the individual sequons. J. Biol. Chem. 263, 6978–6985 [PubMed] [Google Scholar]
- Reddy A., Gibbs B. S., Liu Y. L., Coward J. K., Changchien L. M., Maley F. (1999). Glycosylation of the overlapping sequons in yeast external invertase: effect of amino acid variation on site selectivity in vivo and in vitro. Glycobiology 9, 547–555 10.1093/glycob/9.6.547 [DOI] [PubMed] [Google Scholar]
- Roboti P., High S. (2012). The oligosaccharyltransferase subunits OST48, DAD1 and KCP2 function as ubiquitous and selective modulators of mammalian N-glycosylation. J. Cell Sci. 125, 3474–3484 10.1242/jcs.103952 [DOI] [PubMed] [Google Scholar]
- Ruiz-Canada C., Kelleher D. J., Gilmore R. (2009). Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136, 272–283 10.1016/j.cell.2008.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez L. M., Chirino A. J., Bjorkman P. (1999). Crystal structure of human ZAG, a fat-depleting factor related to MHC molecules. Science 283, 1914–1919 10.1126/science.283.5409.1914 [DOI] [PubMed] [Google Scholar]
- Shakin-Eshleman S. H., Spitalnik S. L., Kasturi L. (1996). The amino acid at the X position of an Asn-X-Ser sequon is an important determinant of N-linked core-glycosylation efficiency. J. Biol. Chem. 271, 6363–6366 10.1074/jbc.271.11.6363 [DOI] [PubMed] [Google Scholar]
- Shibatani T., David L. L., McCormack A. L., Frueh K., Skach W. R. (2005). Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry 44, 5982–5992 10.1021/bi047328f [DOI] [PubMed] [Google Scholar]
- Shrimal S., Ng B. G., Losfeld M-E., Gilmore R., Freeze H. H. (2013a). Mutations in STT3A and STT3B cause two congenital disorders of glycosylation. Hum. Mol. Genet [Epub ahead of print] doi:10.1093/hmg/ddt312 10.1093/hmg/ddt312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimal S., Trueman S. F., Gilmore R. (2013b). Extreme C-terminal sites are posttranslocationally glycosylated by the STT3B isoform of the OST. J. Cell Biol. 201, 81–95 10.1083/jcb.201301031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker M., Linton D., Hitchen P. G., Nita-Lazar M., Haslam S. M., North S. J., Panico M., Morris H. R., Dell A., Wren B. W. et al. (2002). N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298, 1790–1793 10.1126/science.298.5599.1790 [DOI] [PubMed] [Google Scholar]
- Whelan S. A., Lu M., He J., Yan W., Saxton R. E., Faull K. F., Whitelegge J. P., Chang H. R. (2009). Mass spectrometry (LC-MS/MS) site-mapping of N-glycosylated membrane proteins for breast cancer biomarkers. J. Proteome Res. 8, 4151–4160 10.1021/pr900322g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. M., High S. (2007). Ribophorin I acts as a substrate-specific facilitator of N-glycosylation. J. Cell Sci. 120, 648–657 10.1242/jcs.000729 [DOI] [PubMed] [Google Scholar]
- Yan Q., Lennarz W. J. (2002). Studies on the function of oligosaccharyl transferase subunits. Stt3p is directly involved in the glycosylation process. J. Biol. Chem. 277, 47692–47700 10.1074/jbc.M208136200 [DOI] [PubMed] [Google Scholar]
- Zielinska D. F., Gnad F., Wiśniewski J. R., Mann M. (2010). Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 10.1016/j.cell.2010.04.012 [DOI] [PubMed] [Google Scholar]
- Zielinska D. F., Gnad F., Schropp K., Wiśniewski J. R., Mann M. (2012). Mapping N-glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common core machinery. Mol. Cell 46, 542–548 10.1016/j.molcel.2012.04.031 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.