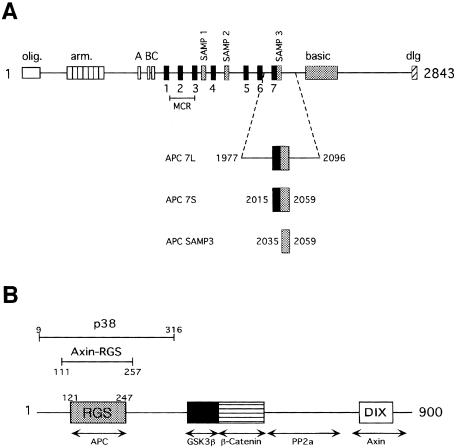
Fig. 1. Primary structure of the Axin and APC proteins. (A) Schematic of APC primary structure. The conserved oligomerization (olig.), armadillo repeat (arm.), basic and discs large interaction (dlg) domains are indicated. The 15 amino acid β-catenin-binding repeats are labeled A, B and C (white boxes). The 20 amino acid β-catenin-binding repeats are labeled 1–7 (black boxes). The Axin-binding repeats are labeled SAMP1–3 (gray boxes). Truncations in the midpoint cluster region (MCR) account for >60% of oncogenic mutations in APC (Miyoshi et al., 1992). The APC 7L, APC 7S and APC SAMP3 constructs used for binding assays are shown below the schematic and their boundaries are indicated. (B) Schematic of Axin primary structure showing regions identified by deletion experiments to be important for protein–protein interactions with APC, GSK3β, β-catenin and protein phosphatase 2a (PP2a). A region of homology to the DIX domain of Dishevelled has been implicated in Axin homodimerization. The region of sequence homology to RGS proteins is indicated. Bars indicate the regions corresponding to the thrombin-defined p38 fragment and the elastase-defined Axin-RGS fragment.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.