Figure 1.
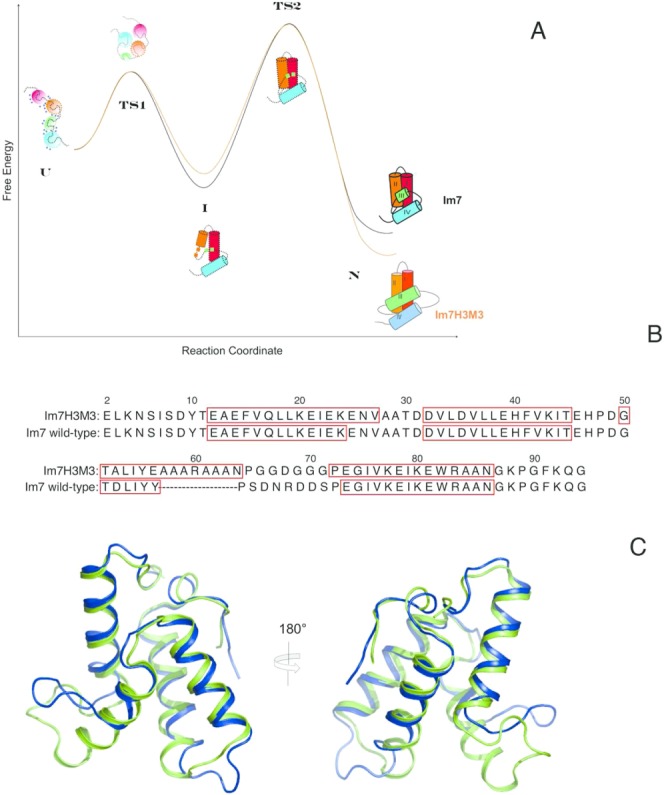
(A) Schematic comparison of the energetics of folding of Im7 and Im7H3M3 based on the thermodynamic and kinetic parameters reported by Knowling et al.24 for Im7H3M3. Except for the cartoon of the native structure of folded Im7H3M3 all other structure cartoons represent Im7 along the folding path. The first is the urea-unfolded state of Im7 which is extended, contains four independent clusters of residues with each cluster containing interacting hydrophobic side chains and has urea molecules (blue circles) associated with it. In the absence of urea the unfolded state (U) rapidly collapses towards the native state (N), traversing transition state 1 (TS1), which lies between the extended (U) and intermediate (I) states, and transition state 2 (TS2) which lies between I and N. (TS1) is almost devoid of secondary structure and lacks a stable hydrophobic core whilst TS2 is a three-helix bundle. The four helices of the native structures are coloured differently, with the important helix III coloured green. Even though helix III has been elongated in Im7H3M3 this protein folds via an on-pathway intermediate24 similarly to wild-type Im7 demonstrating that the formation of an intermediate is an integral feature of the folding mechanism of Im7 that does not result from the short length and low helical propensity of the native helix III. Overall, the elongation of helix III of Im7 to create Im7H3M3 has marginally destabilized the intermediate state by less than 2.5 kJ mol−1 and stabilized the native state by the same amount.24 (B) The Im7 and Im7H3M3 amino acid sequences. The redesigned helix III in Im7H3M3 is longer than its counterpart in Im7. Residues in boxes correspond to α-helices in the native structures. (C) Overlay of the most representative conformer (closest to the average model judged by global RMSDs) from the NMR solution structure ensemble of Im7H3M3 (green)24 and the X-ray crystal structure of Im7 (blue).14 The NMR ensemble of 30 conformers of Im7H3M3 was superimposed onto the crystal structure of Im7 using the SuperPose web server25 global RMSDs over all residues 1.3 Å, heavy atoms 1.1 Å, and backbone atoms 0.8 Å, respectively. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
