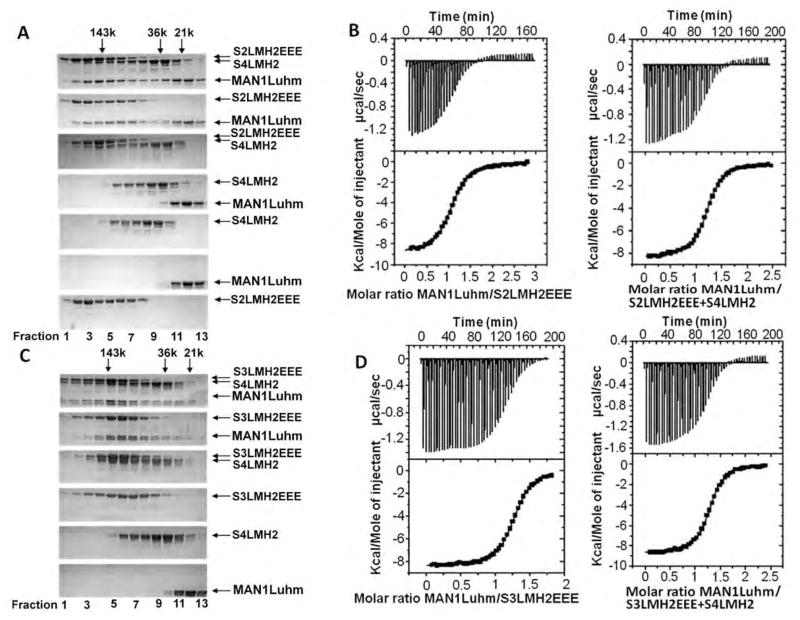
Fig. 5.
The MAN1 fragment interacts with phosphomimetic Smad2 or Smad3 fragments complexed to a Smad4 fragment. (A) Size exclusion chromatography analysis of the interaction between MAN1Luhm (amino acids 755 to 911) and/or a phosphomimetic triple mutant of the Smad2 MH2 domain (S2LMH2EEE) and/or the Smad 4 MH2 domain (S4LMH2). N=2 independent biological replicates. (B) Representative binding curves obtained by ITC when MAN1Luhm was added to S2LMH2EEE,alone or in complex with S4LMH2. Fitting these curves yielded the Kd values in Table 1. (C) Size exclusion chromatography analysis of the interaction between MAN1Luhm and/or a phosphomimetic triple mutant of the Smad3 MH2 domain (S3LMH2EEE) and/or the MH2 of Smad4 (S4LMH2). Eluted fractions were separated on a denaturing gel and stained with Coomassie blue. N=2 independent biological replicates. (D) Representative binding curves obtained by ITC when MAN1Luhm at was added to S3LMH2EEE,alone or in complex with S4LMH2. Fitting these curves yielded the Kd values in Table 1.