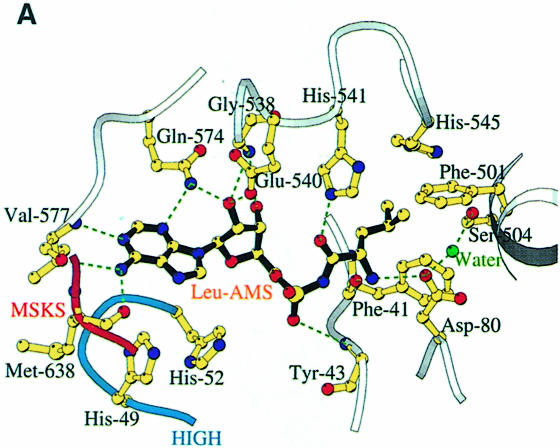
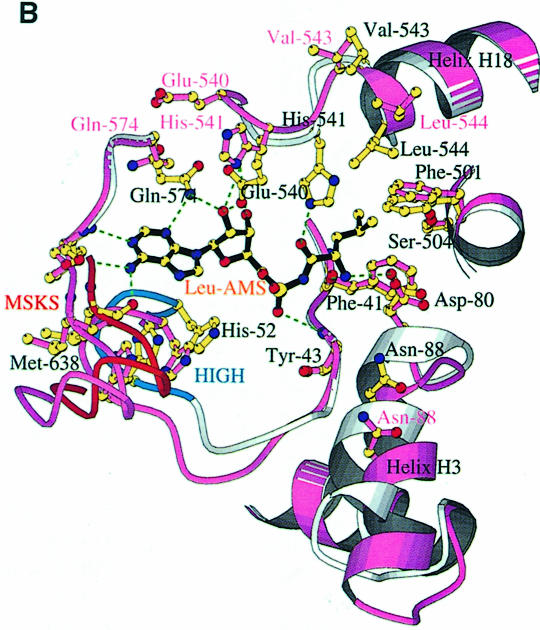
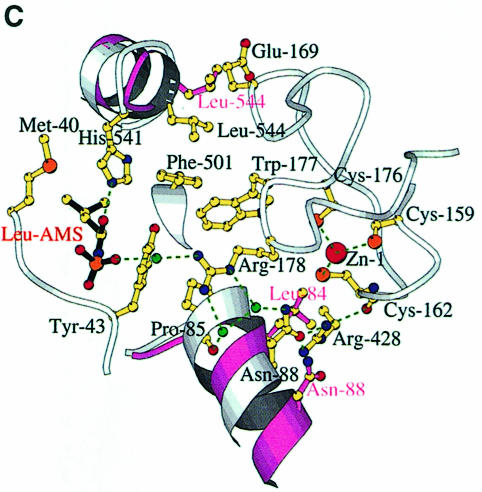
Fig. 4. (A) The LeuAMS binding site showing the major interacting residues. Hydrogen bonds are shown as dashed green lines and a tightly bound water as a green sphere. The catalytically essential class 1 motifs H49IGH and M638SKS are shown in cyan and red, respectively. The side chain of Tyr43 is omitted for clarity, but is visible in (C). (B) The conformational changes associated with LeuAMS binding. The view is the same as in (A). The pink ribbon diagram, pink side chains and pink labels correspond to the apo-structure (mercury derivative) and the grey ribbon and yellow side chains belong to the LeuAMS-bound structure. Upon binding of the adenosine moiety, the HIGH and MSKS loops towards the active centre, Gln574 and Glu540 move to bind the ribose tightly, and helices H18 and H3 refold to permit packing of the ZN-1 domain close to the active site [see the text and (C)]. A sulfate ion (not shown) is bound to His49 and His52 in the apo-structure, but not in the LeuAMS-bound structure. (C) Proximity of Arg178 to the active center in the LeuAMS complex. Colouring as in (B) with water molecules as green spheres and the Zn-1 atom as a red sphere. One of the zinc ligands (His179) and the adenosine moiety of the LeuAMS are omitted for clarity. The positions of Leu544 and Leu84 sterically prevent the packing of the ZN-1 domain close to the active site in the apo-structure.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.