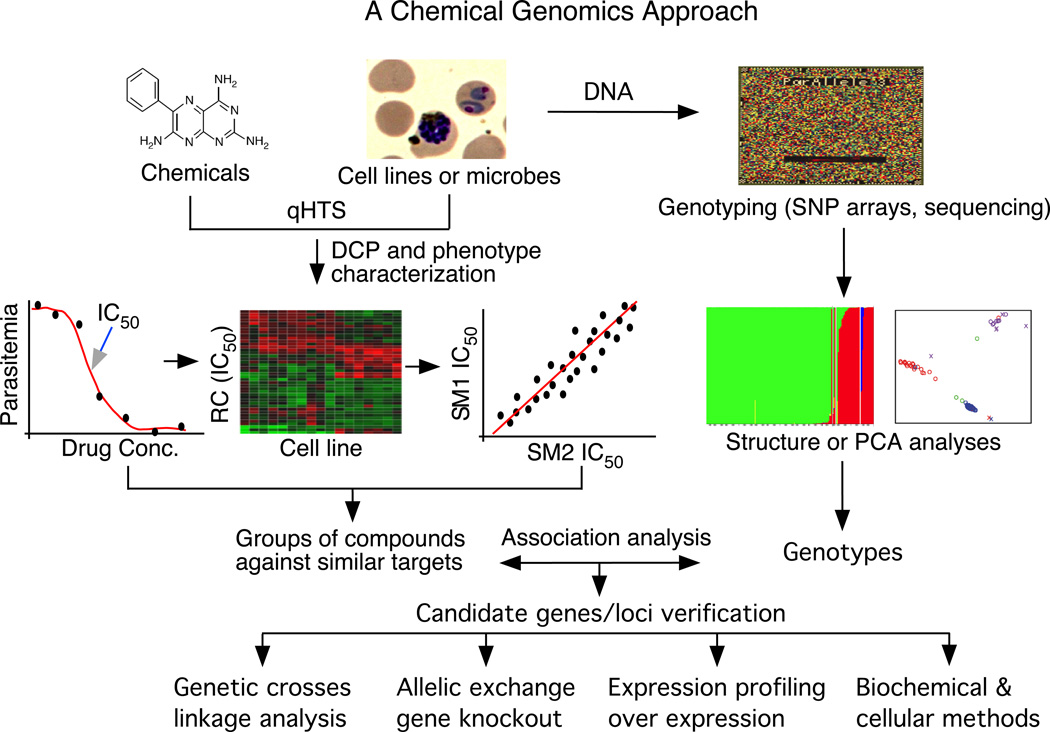
Figure 2.
A chemical genomics approach to identify targets of small molecules. Cell lines or microorganisms are screened against chemical libraries for differences in their responses to the chemicals (differential chemical phenotypes or DCPs). The compounds can be clustered based on the similarity of response patterns (or positive/negative correlation between pairs of compounds) from a panel of cell lines or isolates. Similar response patterns among compounds may suggest that they act on the same or similar targets. DNA samples from the cell lines or isolates are typed using high-throughput genotyping methods such as microarrays or parallel sequencing. Genotypes obtained are analyzed for potential population structures before association analysis. Candidate loci or genes can be further characterized using linkage analysis if progeny from genetic crosses are available. The candidate genes can also be genetically modified or overexpressed to evaluate phenotypic changes. Gene functions can be deduced after additional biochemical and cellular characterizations. Abbreviations: qHTS, quantitative high-throughput screening; RC, response to chemicals; SM1 and SM2, small molecules 1 and 2; SNP, single nucleotide polymorphism; PCA, principal component analysis.