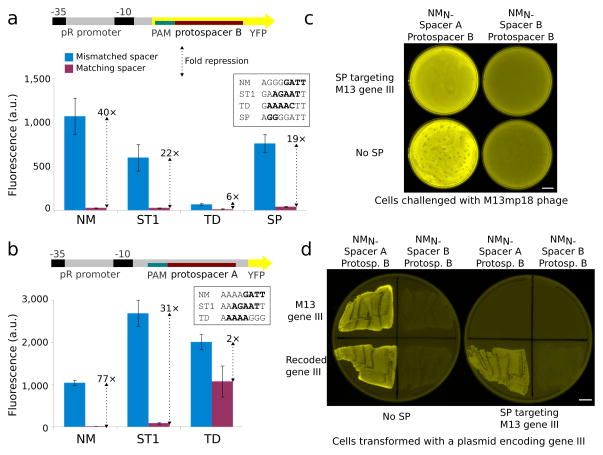
Fig. 4.
Transcriptional repression simultaneous with nuclease activity in bacteria. (a) Reporter plasmids for quantification of Cas9 repression contained protospacer B and a suitable PAM in the non-template strand after the YFP start codon. Normalized cellular fluorescence is shown for mismatched and matched spacer-protospacer pairs. PAMs for each Cas9 are shown. (b) Cas9 ortholog repression was verified with a second reporter plasmid containing protospacer A and a PAM in the non-template strand within the 5′ UTR. Error bars represent the standard deviation of eight independently picked cultures for all repression experiments. (c) Cells containing the plasmids used for NM-mediated repression (Fig. 3a) were transformed with a compatible plasmid encoding SP, its tracrRNA, and a 5-spacer CRISPR locus designed to cleave filamentous phage gene III at multiple sites and challenged with M13mp18. The phage defense plasmid completely prevented plaque formation while preserving YFP repression. (d) Cells were transformed with a compatible plasmid encoding carbenicillin resistance and either wild-type gene III or a recoded version lacking protospacers and plated. Plasmids encoding wild-type gene III were perfectly excluded from cells encoding the SP phage defense, which did not interfere with YFP repression. Scale bars, 10 mm.