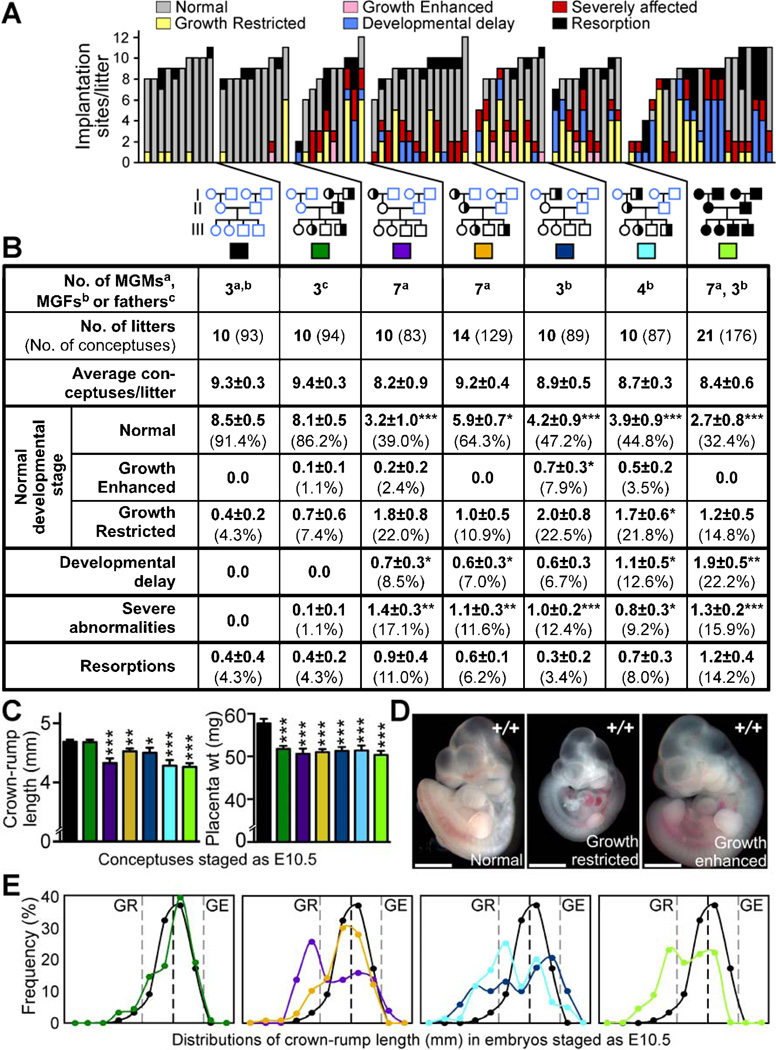
Figure 3. Maternal grandparental Mtrr genotypes are important for a successful pregnancy.
(A-B) Phenotypic frequencies caused by Mtrr+/gt maternal grandmothers (MGMs) or grandfathers (MGFs) at E10.5 compared to C57Bl/6 controls. Mtrr+/gt paternal and Mtrrgt/gt pedigrees were also assessed. Data represented as (A) number of conceptuses/litter with a specific phenotype and (B) average number of phenotypically-affected conceptuses/litter (±SEM) followed by the percentage of total conceptuses in brackets. Colored boxes in (B) indicate the pedigrees assessed in (C) and (E). Pedigrees: circle, female; square, male; blue outline, C57Bl/6 mice; black outline, Mtrr mouse line; white fill, Mtrr+/+; half black/half white fill, Mtrr+/gt; black fill, Mtrrgt/gt. Black asterisks, independent comparison of phenotypic frequencies per litter of each pedigree and C57Bl/6 using Mann-Whitney test. (C) Average embryo crown-rump lengths (±SEM) (left) and average placental weights (±SEM) (right) from conceptuses staged as E10.5 (i.e., 30–40 somite pairs) per pedigree (N=51–89 conceptuses). Statistics: comparison to C57Bl/6 using independent t-test. (D) Representative images of normal, growth-restricted (GR) and growth-enhanced (GE) wildtype embryos at E10.5. Scale bars=1mm. (E) Distribution of crown-rump lengths in E10.5-staged embryos (N=51–89 embryos). Black dotted line, mean; grey dotted lines, two SD from mean. *P<0.05, **P<0.01, ***P<0.005. See also Figure S2 and Table S2.