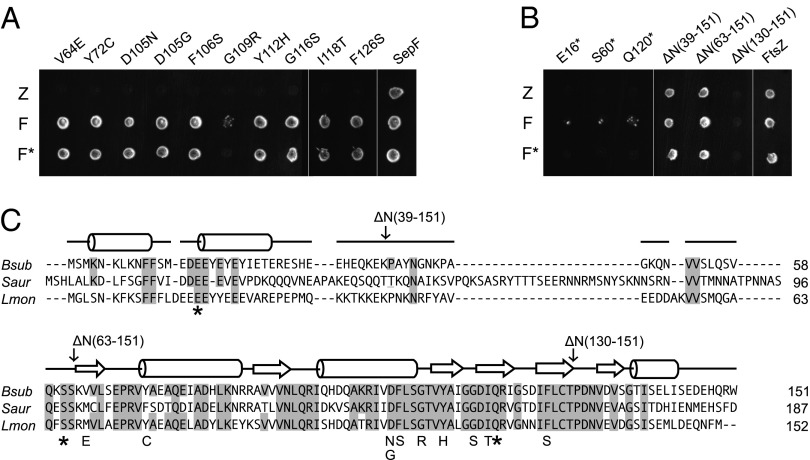
Fig. 1.
Yeast two-hybrid screen for SepF-FtsZ interaction mutants. (A) SepF mutants were created using error-prone PCR, and FtsZ-interaction deficient mutants were selected using the yeast two-hybrid assay. Diploid colonies were obtained by mating PJ69-4α strains harboring pGBT9 derivatives containing sepF mutants, and wild-type sepF, with PJ69-4A strains that harbored pGAD424 derivatives containing ftsZ (Z), wild-type sepF (F), or the corresponding sepF mutants (F*). (B) Yeast two-hybrid interactions of the nonsense mutants and the different N-terminal deletion constructs. Remaining amino acid regions are indicated between parentheses. A and B are discontinuous images of the same plate. (C) Amino acid sequence alignment of SepF proteins, from B. subtilis (Bsub), Staphylococcus aureus (Saur), and Listeria monocytogenes (Lmon), using ClustalW. Secondary structure prediction (using PSIPRED) for B. subtilis SepF is shown above the sequences, whereby α-helices and β-sheets are indicated by rods and arrows, respectively. Conserved amino acids are shaded. Yeast two-hybrid mutations are indicated.