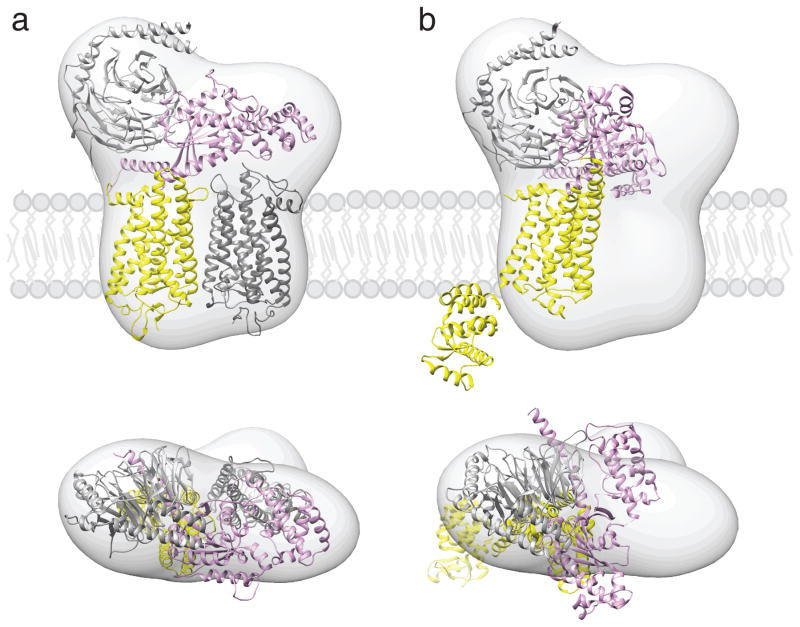
Fig. 3.
Proposed location of GPCR-G protein complexes in a phospholipid bilayer. Semi-empirical model of the complex formed between light-activated rhodopsin dimer and a Gt heterotrimer (a) (Jastrzebska et al. 2011) and a structure of T4L-β2AR-Gs complex (b) (Rasmussen et al. 2011b) were fitted into a 3D molecular map derived from EM-single particle reconstructions of negatively stained, crosslinked rhodopsin-Gt complexes purified in LMNG. Best fitting of the rhodopsin-Gt model was achieved for rhodopsin dimer and Gt hereotrimer (GOT1) after 30° hinge-like motion of the α-helical domain of Gt was applied. Fitting the T4L-β2AR-Gs-nanobody structure into our EM 3D map leaves enough unoccupied space to accommodate a second molecule of this receptor, conformation of Gsα helical domain is inconsistent with our EM-density. The photoactivated rhodopsin that binds C-terminus of Gtα and molecule of T4L-β2AR is depicted with yellow. The second rhodopsin molecule is shown in dark grey. Gα is colored pink; Gβ and Gγ are colored light grey.