Abstract
The gene nhr-6 encodes the Caenorhabditis elegans ortholog of the NR4A nuclear receptor. We determined the biological functions of NHR-6 through the isolation and characterization of a deletion allele of nhr-6, lg6001. We demonstrate that nhr-6 has an essential role in the development of the C. elegans somatic gonad. Specifically, nhr-6 is required for the development of the hermaphrodite spermatheca, a somatic gonad organ that serves as the site of sperm storage and oocyte fertilization. Using a variety of spermatheca cell markers, we have determined that loss of nhr-6 function causes severe morphological defects in the spermatheca and associated spermathecal valves. This appears to be due to specific requirements for nhr-6 in regulating cell proliferation and cell differentiation during development of these structures. The improper development of these structures in nhr-6(lg6001) mutants leads to defects in ovulation and significantly reduced fecundity of C. elegans hermaphrodites. The phenotypes of nhr-6(lg6001) mutants are consistent with a role for nhr-6 in organogenesis, similar to the functions of its mammalian homologs.
Keywords: nhr-6, NR4A, C. elegans, spermatheca, spermatheca-uterine valve
INTRODUCTION
The formation of organs requires the precise regulation and coordination of cell proliferation, differentiation, morphogenesis and growth (Sears and Nevins, 2002; Vidwans and Su, 2001). The nematode Caenorhabditis elegans is an outstanding model for the genetic dissection of organ formation. The invariant cell lineage allows for the analysis of organ formation to be performed at the single-cell level which, in turn, provides a framework for the careful dissection of genetic processes regulating proliferation and differentiation of various organ lineages (Kipreos, 2005).
An adult C. elegans hermaphrodite consists of only 959 somatic cells (Kimble and Hirsh, 1979; Sulston and Horvitz, 1977). Nonetheless, C. elegans possesses several complex organ systems. For example, the egg-laying machinery of C. elegans requires the coordinated development of two organs, the vulva and uterus. The genes that have been identified in the genetic analysis of these organ systems have provided insight into the cell-cell signaling processes that regulate cell division patterns, cell differentiation, morphogenesis, and the development of the physical interactions between these two interconnected organs (Newman et al., 1999; Newman et al., 2000; Palmer et al., 2002; Rajakumar and Chamberlin, 2006; Wang and Sternberg, 2001).
Despite this progress, much remains to be understood about the organ formation process. One area of interest is the role of transcription factors in coordinating lineage-specific patterns of cell division and differentiation. Transcription factors that function during organogenesis must have the ability to respond to and integrate multiple signaling pathways to control cell proliferation and cell differentiation. One group of transcription factors that have this ability are the nuclear receptors (NRs), among the most important regulators of gene expression in the metazoa. NRs are a diverse group of transcription factors that are unique in their ability to regulate the expression of specific genes in response to a wide variety of intercellular and intracellular signals (Mangelsdorf et al., 1995). The most intensively studied and well understood NRs are those that function as receptors for signaling molecules such as endocrine hormones and cellular metabolites. In addition to the regulation of core cell physiological processes, NRs also function as key transcriptional regulators during development. Common developmental functions of NRs include early embryonic patterning, cell fate specification, organogenesis and invertebrate life cycle progression (Giguere, 1999; King-Jones and Thummel, 2005).
One particular group of NRs, NR4A, has recently emerged as developmentally important transcriptional regulators. Structural studies have demonstrated that NR4A NRs are not ligand regulated in the canonical NR fashion (Wang et al., 2003). Therefore, it is likely that NR4A NRs are true unliganded (orphan) receptors that mediate cellular processes through interactions with signal transduction pathways (Maira et al., 2003; Pekarsky et al., 2001; Swanson et al., 1999; Wang et al., 2003; Wingate et al., 2006). The three mammalian paralogs of the NR4A subgroup (NR4A1, NR4A2, and NR4A3) have been implicated in a vast array of biological processes including immune system development, metabolism, inflammation, atherogenesis, steroidogenesis, central nervous system development, and organogenesis (Maxwell and Muscat, 2006). Despite their biological importance, much remains to be learned regarding the precise functions and cellular regulation of these transcription factors. However, organogenesis, tissue differentiation, and cell proliferation in multiple contexts appear to be important biological functions of the NR4A subgroup (Castro et al., 2001; Kolluri et al., 2003; Martinez-Gonzalez et al., 2003; Suzuki et al., 2003; Zeng et al., 2006). As examples of this, Nurr1 (NR4A2) has been established to be a critical differentiation factor for mid-brain dopaminergic neurons (Zetterstrom et al., 1997) and NOR-1 (NR4A3) is required for the proliferation and differentiation of inner ear epithelia, as well as hippocampal and pyramidal cell development (Ponnio et al., 2002; Ponnio and Conneely, 2004).
Here we characterize the developmental functions of nhr-6, the sole NR4A homolog encoded in the C. elegans genome (Sluder and Maina, 2001). We present evidence that nhr-6 is important for reproduction in C. elegans and is required for proper morphogenesis of the spermatheca, a component of the C. elegans somatic gonad involved in oocyte ovulation and fertilization. We also demonstrate that nhr-6 has specific functions in cell proliferation and cell differentiation during development of the spermatheca and spermatheca-uterine valve. This work establishes nhr-6 as a model system for investigating the cellular activities of NR4A in the regulation of gene expression during development, and also establishes the spermatheca as an additional C. elegans system for investigating the genetic regulation of organogenesis.
MATERIALS AND METHODS
Nematode strains
C. elegans was cultured using standard protocols (Brenner, 1974). The following strains were utilized in this study: N2 (Bristol); XY1004 (nhr-6(lg6001)); GR1371 (eri-1(mg366)); JJ532 (pie-1(zu154) unc-25(e156)/qC1 dpy-19(e1259) glp-1(q339)); SU93 (jcIs1[ajm-1::GFP + rol-6(su1006)]); DG1576 (tnIs5[lim-7::GFP + rol-6(su1006)]); PS3662 (syIs63[cog-1::GFP + unc-119(+)]); UA2 (baEx2[lis-1::GFP + rol-6(su1006)]); HR596 (mel-11::GFP); HR606 (let-502::GFP); MT2405 (ced-3(n717) unc-26(e205); DZ325 (ezIs2[fkh-6::GFP + unc-119(+)]; him-8(e1489); CB3297 (vab-9(e1744); him-5(e1490); NL2099 (rrf-3(pk1426); PS3411 (cog-1(sy607)/mnC1 dpy-10(e128) unc-52(e444); nhr-6(tm870).
The RNAi sensitized fkh-6::GFP expressing strain rrf-3(pk1426); ezIs2 was constructed by crossing rrf-3(pk1426) males to vab-9(e1744); ezIs2 hermaphrodites. vab-9 was utilized as a chromosome II marker. GFP expressing non-Vab F2 animals were isolated. Homozygous rrf-3(pk1426); ezIs2 lines were confirmed by GFP expression analysis and sterility at 25°C (which confirmed the rrf-3(pk1426) background).
The cog-1(sy607); nbIs1000 strain was generated by crossing cog-1(sy607)/+ males to nbIs1000 hermaphrodites. F2 Cog Rol animals were isolated and a homozygous cog-1(sy607); nbIs1000 strain was established. cog-1(sy607) homozygotes exhibit extremely low brood sizes but they can be maintained as a homozygous line. This strain was further confirmed by PCR verification of the cog-1(607) deletion (Palmer et al., 2002).
Isolation of the lg6001 deletion allele
A deletion mutant of nhr-6 was isolated by PCR screening of mutant libraries representing approximately 4,500,000 haploid genomes. Libraries were constructed by trimethylpsoralen treatment in combination with UV irradiation of wild-type N2 animals (Jansen et al., 1997). Subpools were screened for a deletion in the nhr-6 gene by nested PCR. The first round was performed using primers EG60.1 (5′-TGCTACTCCGCCTTCTCAAT-3′) and EG60.2 (5′-GCTCGATCATTGCACACA-3′), the second round with primers EG60.3 (5′-GACCGCTCATTAATCGGATGCG-3′) and EG60.4 (5′-AGGTTGGTGTGGAGAGGTTG-3′). The isolated nhr-6(lg6001) deletion strain (XY1004) was found to be homozygous viable and was outcrossed to N2 five times.
Reverse Transcription-Polymerase Chain Reaction and Western analysis
To identify full-length, SL1 trans-spliced nhr-6 mRNA isoforms 2 μg of total RNA from mixed-stage N2 cultures was reverse-transcribed using the Protoscript First Strand cDNA Synthesis kit (New England Biolabs) with oligo-dT primer. Primary PCR was performed using 1 μl of cDNA template and a primer corresponding to the SL1 trans-splice leader sequence and an nhr-6 3′UTR specific primer (5′TTGATATTCGGGGATTGGAA-3′). Secondary PCR was performed using the SL1 primer and a nested nhr-6 3′UTR primer (5′-GGGGATTGGAATTCACACAA-3′). PCR products were gel purified and sequenced.
Chicken anti-NHR-6 antibodies were generated by Covance Research Products (Denver, PA) using purified His-tagged NHR-6/LBD antigen. The antibody was affinity purified against the antigen followed by affinity purification against an MBP fusion of the NHR6/LBD. SDS-PAGE was performed using equal amounts of protein from mixed-staged N2 and nhr-6(lg6001) nematode cultures. Western blots were probed with primary antibodies followed by HRP-linked anti-chicken secondary antibodies. Blots were washed in Tris-buffered saline containing 0.3% Tween 20 and 0.5% Triton-X prior to adding HRP substrate.
Immunolocalization with anti-NHR-6/LBD was attempted but only gave diffuse, apparently non-specific staining in both wild-type and mutant animals. Several approaches and fixation conditions were attempted. The Western analysis described above required very stringent wash conditions to eliminate background signal and this characteristic of the antibody likely affected the immunolocalization experiments.
nhr-6(lg6001) phenotypic analysis
nhr-6(lg6001) homozygotes are viable but adults degenerate rapidly after production of the first few eggs. Therefore, all microscopic phenotypic analyses (except where specifically noted) of nhr-6(lg6001) animals were performed during the young adult stage prior to or soon after the first ovulation.
Construction of an nhr-6 RNAi vector was previously described (Gissendanner et al., 2004). RNAi was performed using a standard RNAi feeding method (Kamath et al., 2001). Robust nhr-6 RNAi requires utilizing a strain with enhanced RNAi sensitivity. In this study we utilized the RNAi sensitive mutant eri-1(mg366) (Kennedy et al., 2004). Synchronized eri-1(mg366) L1 larvae derived from alkaline hypochlorite egg preparation were placed on lawns of HT115 E. coli bacteria induced with 1mM IPTG to express nhr-6 double-stranded RNA. Nematodes were grown on induced bacteria at 20°C until they reached the young adult stage.
Brood count experiments were performed by placing single young adults on brood count plates (plates that contain a narrow strip of OP50 E. coli bacteria). Animals were moved every 12-24 hours until egg production stopped or until animals “bagged”, as was frequently observed with nhr-6(lg6001) mutants. Each time animals were moved, eggs and larvae were counted. 24 hours after adults were moved, each plate was scored for arrested eggs and viable larvae. The viable larvae counts are reported in Table 1. To assess egg morphology and lethality specifically, groups of young adult animals (see Table 2) were placed on a brood count plate and eggs were collected for 24 hours. Adult animals were removed from the plate after egg collection. Abnormal egg morphology was noted when eggs exhibited small size, round shape or other irregular shapes compared to the normal oval shaped eggs produced by wild-type nematodes. Embryonic lethality was assessed 24 hours after egg collection.
Table 1.
Reproductive phenotypes in nhr-6 loss-of-function animals
| Genotype | N (adults scored) |
Avg. Viable Brood Size |
Range |
|---|---|---|---|
| +/+ | 3 | 230 ± 32 | 205-266 |
| nhr-6(lg6001)/nhr-6(lg6001) | 35 | 8 ± 101 | 0 - 38 |
| nhr-6(lg6001)/nhr-6(tm870) | 14 | 4 ± 7 | 0- 21 |
|
nhr-6(lg6001)/nhr-
6(lg6001); nbEx1000 |
5 | 182 ± 21 | 159-205 |
|
nhr-6(lg6001)/nhr-
6(lg6001); nbEx1010 |
5 | 163 ± 45 | 105-222 |
|
nhr-6(lg6001)/qC1 dpy-
19(e1259) glp-1(q339) |
3 | 228 ± 34 | 194-261 |
| eri-1(mg366); nhr-6(RNAi)2 | 8 | 5 ± 5 | 2-16 |
| eri-1(mg366); GFP(RNAi) | 4 | 185 ± 24 | 166-220 |
Brood size counts include progeny from hermaphrodites that bagged without laying eggs.
It had been previously observed that nhr-6 RNAi experiments in a wild-type background yield similar phenotypes. However, the phenotypes are less severe and incompletely penetrant (Gissendanner et al., 2004). This indicates that nhr-6 is somewhat refractory to RNAi in a wild-type background.
Table 2.
Abnormal Egg Morphology in nhr-6 loss-of-function animals
| Genotype | N (eggs scored) |
% abnormal morphology |
% arrest |
|---|---|---|---|
| +/+ | 99 | 0 | 0 |
|
nhr-6(lg6001)/nhr-
6(lg6001) |
152 | 78 | 341 |
|
nhr-6(lg6001)/nhr-
6(lg6001); nbEx1000 |
66 | 0 | 3 |
|
nhr-6(lg6001)/nhr-
6(lg6001); nbEx1010 |
93 | 7.9 | 0 |
|
nhr-6(lg6001)/qC1 dpy-
19(e1259) glp-1(q339) |
238 | 0 | 0 |
| eri-1(mg366); nhr-6(RNAi) | 224 | 59 | 391 |
| eri-1(mg366); GFP(RNAi) | 135 | 0 | 0 |
Eggs scored were collected from 5 adult hermaphrodites (+/+, nhr-6(lg6001)/nhr-6(lg6001); nbEx1000, nhr-6(lg6001)/nhr-6(lg6001); nbEx1010, nhr-6(lg6001)/qC1, eri-1(mg366); GFP(RNAi)), 25 adult hermaphrodites (eri-1(mg366); nhr-6(RNAi), or 40 adult hermaphrodites (nhr-6(lg6001)/nhr-6(lg6001) during a 24 hour lay period. Embryonic arrest was assessed 24 hours after the end of the lay period.
All arrested eggs displayed abnormal egg morphology.
To assess the mating ability of nhr-6(lg6001) males, three spontaneously arising young adult nhr-6(lg6001) males were removed from a mixed stage culture and mated to 3 dpy-17(e164) unc-32 (e189) young adult hermaphrodites. >95% of the F1 progeny were cross progeny.
Phalloidin-staining and myosin heavy chain A (MHCA) and MH27 immunohistochemistry was performed on gonads extruded from young adults. Gonads were extruded in PBS/0.01% tetramisole using 25G needles and collected in 1.5 ml siliconized tubes. Fixation of extruded gonads and subsequent staining using anti-MHCA antibody (monoclonal antibody 5.6, gift from David Miller) were performed exactly as previously described (Rose et al., 1997). For anti-AJM-1 staining, the monoclonal MH27 antibody (Developmental Studies Hybridoma Bank) was used at 1:50 dilution. For phalloidin-staining, extruded gonads were fixed in 1.25% paraformaldehyde for 10min at room temperature and washed twice in PBS/0.5% Triton-X (PBST). Gonads were stained with 0.165 μM FITC-Phalloidin (Sigma-Aldrich) and 1 μg/ml DAPI for one hour in the dark and then washed twice in PBST.
Construction and analysis of nhr-6(lg6001); fog-2(q71) double mutants
nhr-6(lg6001); fog-2(q71) double mutants were constructed by crossing fog-2(q71) males to IP1005 (nhr-6(lg6001); jcIs1) hermaphrodites. IP1005 was utilized so that spermatheca morphology could be assessed in the double mutants using the AJM-1::GFP marker. L4 F2 progeny from the cross were singled to a large number of NGM plates. Both Rol (indicating presence of jcIs1) and non-Rol F2 progeny were selected. Plates were screened for sterile animals the following day. Sterile animals were observed with high magnification to confirm Fog phenotype (absence of sperm and fertilized eggs) and scored for ovulation defects. 34 Fog animals were observed. 12/34 contained a proximal mass of oocytes, consistent with ovulation in the absence of sperm. Single animals with and without ovulation defects were removed and genotyped for the lg6001 allele by PCR using primers 5′-TGCTCTTTTGTCGCAGCTAA-3′ and 5′-GTTACAAGGCTTCCCCGCTA-3′, which detects both the wild-type and lg6001 deletion allele. All animals with ovulation defects (ovulation in the absence of sperm associated with a proximal mass of fragmented oocytes and lack of oocyte compaction in the gonad arm) were found to be homozygous for the nhr-6(lg6001) allele. Normal Fog animals were either +/+ or nhr-6(lg6001)/+.
Reporter gene analysis
Construction and initial analysis of the full-length nhr-6::GFP reporter (pCG52) has previously been described (Gissendanner et al., 2004). The pCG52-bearing extrachromosomal array, nbEx1002, was integrated using a methylmethane sulfonate-based integration protocol graciously provided by Yinhua Zhang, New England Biolabs (manuscript in preparation). One integrated line (nbIs1000) was isolated and this line was outcrossed to N2 three times to generate strain IP1018. This strain exhibited an expression pattern identical to the nbEx1002 strain, IP1004.
Transcriptional reporter constructs for the three putative regulatory regions (R1-3, corresponding to constructs pCG53-55, see Figure 3) were constructed by PCR amplification of the specified genomic regions using a previously described nhr-6 genomic clone as template (Gissendanner et al., 2004). Amplified products were cloned into the PstI/XmaI (for pCG53 and pCG54) or SphI/BamHI (for pCG55) sites of pPD95.67 (gift from Andy Fire). The following primers were utilized: R1 (5′-CCCAATGATCCGATCTGTTT-3′ and 5′-CACAGCAAAAGCATTGTG-3′); R2 (5′-AGCTGTTCCGGAAGTAGTAAGC-3′ and 5′-CTGAAAATGTTCAGAATTTTAACG-3′); R3 (5′-GTGAGCTCTTTTTCCAAGAAC-3′ and 5′-CTGCAAACTTTCAATTTCAGAC-3′).
Figure 3. Ovulation in nhr-6(lg6001); fog-2(q71) females.
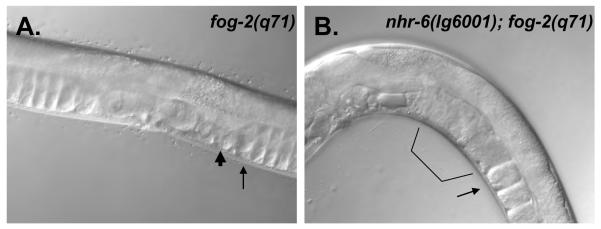
DIC micrographs of fog- 2(q71) (A) and nhr-6(lg6001); fog-2(q71) (B) adults. The spermatheca (thick arrow) and proximal oocyte (thin arrow) are indicated for the animal in (A). Note compaction of the oocytes in the gonad arm (arrow). The animal in (B) lacks the oocyte compaction (arrow) and the proximal gonad is filled with a mass of oocytes and oocyte fragments (bracket).
Transgenic nematodes were generated by injecting 10 μg/ml reporter and 100 μg/ml pRF4 (Mello and Fire, 1995) for pCG53 and pCG54 and 20 μg/ml reporter and 100 μg/ml pRF4 for pCG55. Two independent transgenic lines were analyzed for each construct. Expression from pCG53-bearing arrays was never observed at any stage of development.
Construction of strains bearing nhr-6(lg6001) and nhr-6(tm870)
The following nhr-6(lg6001) strains were constructed for this study: IP1001 (nhr-6(lg6001)/ qC1 dpy-19(e1259) glp-1(q339)); IP1005 (nhr-6(lg6001); jcIs1[ajm-1::GFP::GFP + rol-6(su1006)]); IP1014 (nhr-6(lg6001); tnIs5[lim-7::GFP + rol-6(su1006)]); DN2 (nhr-6(lg6001); syIs63[cog-1::GFP + unc-119(+)]); DN6 (nhr-6(lg6001); nbIs1000); DN7 (nhr-6(lg6001); baEx2[lis-1::GFP + rol-6(su1006)]); DN10 (nhr-6(lg6001); unc-26(e205)ced-3(n717); nbIs1000); DN18 (nhr-6(lg6001)/+; let-502::GFP); DN19 (nhr-6(lg6001)/+; mel-11::GFP). Due to the reduced fecundity of nhr-6(lg6001) homozygotes, construction of these strains required utilizing large numbers of nhr-6(lg6001) hermaphrodites in the P0 crosses. Typically, 30-40 nhr-6(lg6001) L4 hermaphrodites crossed to 30-40 N2 young adult males would generate sufficient numbers of F1 nhr-6(lg6001)/+ males for subsequent crosses. nhr-6(lg6001) homozygotes are recognized in the F2 generation of these crosses by the presence of large numbers of bagging animals. Strains bearing the nhr-6(lg6001) allele were confirmed by both phenotypic analysis and single worm PCR using primers to detect the nhr-6(lg6001) deletion. For strain DN10, unc-26(e205) was used to track the ced-3(n717) chromosome in the crosses. Three independent lines were isolated and all failed to suppress the nhr-6(lg6001) phenotype.
A balanced nhr-6(tm870)/ qC1 dpy-19(e1259) glp-1(q339) strain was generated by the same strategy for nhr-6(lg6001). nhr-6(lg6001)/ nhr-6(tm870) trans-heterozygotes were generated by crossing nhr-6(lg6001)/qC1 males to nhr-6(tm870)/qC1 hermaphrodites. F1 cross progeny were singled to plates. F1 animals were isolated that exhibited an Nhr-6 phenotype. The brood sizes of these animals was assessed and the parent animals were genotyped by PCR using primers 5′TGCTCTTTTGTCGCAGCTAA-3′ and 5′-AGCTCGATCATTGCACACAG-3′ to distinguish the lg6001 and tm870 deletions in a single reaction. Data were used only from confirmed nhr-6(lg6001)/ nhr-6(tm870) trans-heterozygotes (14/15 animals tested).
Nuclei Counts
Extruded gonads were DAPI stained after fixation as described above. GFP defined cell counts were performed using between 10 and 12 optical sections though the gonad plane on anesthetized IP1018 (chromosomally integrated nhr-6::gfp transgene, nbIs1000) and nhr-6(lg6001); nbIs1000 young adults. Micrographs were taken using a Diagnostic Instruments Spot Cam version 2.3.1.1 (Diagnostic Instruments Inc., MI). Micrographs of both GFP expression and DIC images were generated in 16 bit mono Tiff format and aligned in separate layers using Adobe Photoshop CS2 (Adobe Systems Inc., CA). Areas of GFP expression were defined with an opacity of less than 50%. Nuclei, identified on DIC images within the defined GFP expression area, were marked on a separate layer and counted, excluding those nuclei in the spermathecal valves.
Rescue assays
Rescue constructs bearing wild-type nhr-6 coding sequence were injected into IP1001 hermaphrodites. Stable transgenic lines were isolated and nhr-6(lg6001) homozygotes bearing the rescue arrays were recovered. A previously described nhr-6 genomic clone, pLH1 (Gissendanner et al., 2004), was injected at a concentration of 1 μg/ml along with 100 μg/ml of pRF4 to generate rescuing array nbEx1000. To generate a rescuing construct under control of the R3 (intron 5) promoter, the pLH1 genomic clone was digested with BamHI, end-filled, and then digested with BlpI. This removes all coding and regulatory sequences upstream of intron 5 as well as most of intron 5. The remaining fragment was gel-purified and ligated to a HindIII (end-filled)/BlpI intron 5 fragment from the pCG55 transcriptional reporter construct, thus replacing the intron 5 sequence lost in the first step. The final construct consisted of intron 5 followed by all downstream nhr-6 genomic sequence. The construct was injected at a concentration of 10 μg/ml along with 100 μg/ml pRF4. Two stable arrays were generated and one array, nbEx1010, rescued nhr-6(lg6001) homozygotes.
RESULTS
nhr-6 encodes two protein isoforms and is required for normal reproduction
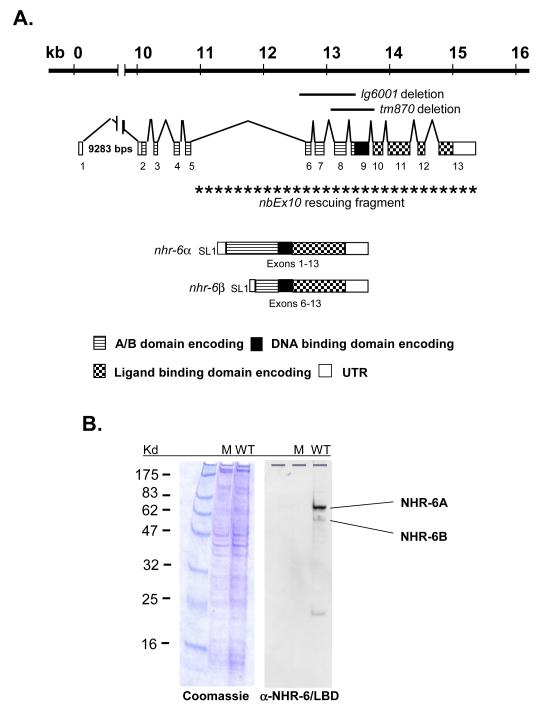
nhr-6 gene structure and phylogenetic placement in the NR4A group has been previously described (Gissendanner et al., 2004; Robinson-Rechavi et al., 2005; Sluder and Maina, 2001). To further characterize the nhr-6 gene, we performed SL1 RT-PCR and determined that nhr-6 encodes two mRNA isoforms (α and β), differentiated by alternative SL1 trans-splicing (Fig. 1A). The shorter nhr-6β lacks the first five exons found in nhr-6α and the 5′ end of the shorter isoform is defined by trans-splicing of SL1 to exon 6. The shorter isoform encodes an NR4A receptor with a shortened N-terminal A/B domain. Western blot analysis (Fig. 1B) of mixed-stage nematodes reveals the presence of two NHR-6 proteins: NHR-6A, and a smaller protein, NHR-6B. The sizes of the two proteins revealed by the Western analysis is consistent with the predicted sizes encoded by the mRNA isoforms (69kDa for NHR-6A and 53kDa for NHR-6B). An ~20kDa protein is also detected in the Western analysis. However, no nhr-6 mRNA has been isolated that could code for a protein of this size. One possibility is that the ~20kd band could represent a proteolytic product of the NHR-6 protein.
Figure 1. nhr-6 gene structure and protein expression.
(A) Structure of the nhr-6 gene. The two mRNA isoforms, α and β, are defined by the alternate SL1 trans-splice sites. The nhr-6β isoform consists of exons 6-13. Exon numbers, the two deletion alleles, and the nhr-6β genomic sequence found in the nbEx10 rescuing array are indicated. (B) Western blot analysis of NHR-6 in wild-type (WT) and nhr-6(lg6001) (M) nematodes. NHR-6A and NHR-6B isoforms are indicated. Coomassie stained gel on the left indicates protein loading.
A deletion allele of nhr-6, lg6001, was isolated using a PCR-based chemical mutagenesis screen. The lg6001 allele deletes the 3′ splice site of intron 5, exons 6-8, and the 5′ end of exon 9 (Fig. 1A). This deletion removes coding sequence common to both mRNA isoforms and Western blot analysis demonstrates that NHR-6A and NHR-6B proteins are not expressed in nhr-6(lg6001) mutant animals, indicating that nhr-6(lg6001) is a molecular null allele (Fig. 1B).
Animals homozygous for nhr-6(lg6001) are viable but exhibit severe reproductive defects (Table 1). A component of this reproductive defect is a highly penetrant “bagging” phenotype, where the failure to properly lay eggs results in internal hatching of larvae. 21/35 nhr-6 (lg6001) homozygous hermaphrodites scored exhibited a “bagging” phenotype. However, homozygous mutants frequently laid eggs prior to bagging. The number of viable offspring from non-bagging nhr-6 (lg6001) homozygous animals ranged from 0-38 (average 7.0 + 12.8). 8/14 non-bagging hermaphrodites failed to produce to viable progeny. Therefore, nhr-6(lg6001) mutants have a specific defect in the ability to generate progeny that is not solely due to an egg-laying defect.
Another phenotype of nhr-6(lg6001) mutants is abnormal egg morphology (Table 2). Wild-type hermaphrodite eggs have a distinct oval shape (Fig. 2A, A’). However, a majority of eggs laid by nhr-6(lg6001) homozygous hermaphrodites exhibit a range of abnormal morphology. Phenotypes ranged from small or round eggs to larger eggs with irregularly shaped borders (Fig. 2B and B’). Of the eggs laid by nhr-6(lg6001) homozygous hermaphrodites (n=152 eggs scored), 78% exhibited an abnormal egg morphology phenotype and 34% arrested development prior to hatching (Table 2). Only the most severely abnormal eggs arrested development. Normally shaped eggs and eggs with less severe abnormal morphology completed embryonic development and the hatched larvae were viable (Fig. B’). The reproductive and egg morphology phenotypes were rescued by transgenes bearing wild-type nhr-6 sequence (Table 1). An additional allele of nhr-6, tm870, isolated by the S. Mitani laboratory (http://www.shigen.nig.ac.jp/c.elegans/mutants/DetailsSearch?lang=english&seq=870), also displays the same phenotypes as lg6001 (data not shown). This allele is an 874 bp deletion that overlaps with lg6001 (Fig. 1A). nhr-6(lg6001)/nhr-6(tm870) trans-heterozygotes display low brood size phenotypes similar to nhr-6(lg6001) homozygotes (Table 1). In addition, the brood size and abnormal egg morphology phenotypes are also observed in nhr-6(RNAi) animals (Table 1 and Table 2). Therefore, we conclude that the observed phenotypes are due specifically to loss of nhr-6 function. Abnormal egg morphology, embryonic arrest and decreased brood size are not observed in nhr-6(lg6001)/+ hermaphrodites, thus, lg6001 appears to be fully recessive (Table 2).
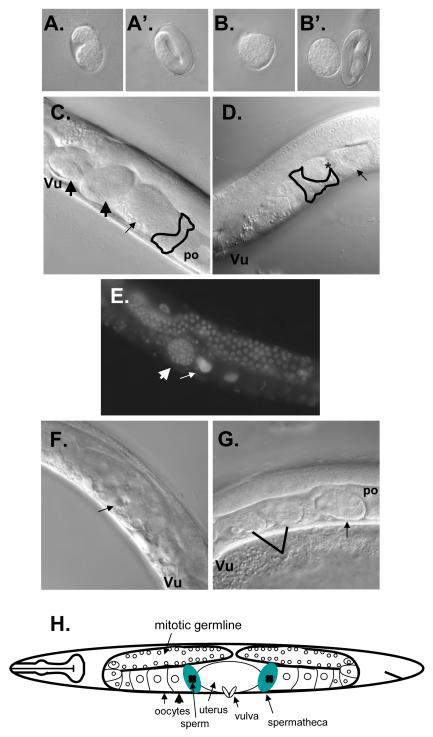
Figure 2. nhr-6(lg6001) phenotypes.
(A, A’) Wild-type egg morphology. (B, B’) Typical abnormal egg morphology of nhr-6(lg6001) mutants. The embryo in (B) has failed to undergo morphogenesis. Note rounded shape compared to wild-type embryo in (A). The embryo to the right in (B’) has developed to the three-fold stage but egg morphology is still abnormal (compare to A’). (C) Wild-type adult hermaphrodite. During ovulation the proximal oocyte (po) is ovulated into the spermatheca (outlined) and is fertilized (thin arrow). Fertilized oocytes then enter the uterus (thick arrows) where they undergo early development prior to being laid through the vulva (Vu). (D) Adult nhr- 6(lg6001) hermaphrodite. In this animal the proximal oocyte (thin arrow) is splitting in two during the ovulation process and contains endomitotic nuclei (*). (E) DAPI staining of an nhr-6(lg6001) hermaphrodite. Thin arrow indicates the increased, diffuse nuclear DNA staining resulting from endomitosis. Thick arrow indicates a well-developed egg residing within the spermatheca. (F) Older nhr-6(lg6001) hermaphrodite displaying gonadal degeneration. (G) nhr-6 (lg6001) adult hermaphrodite with misplaced developing embryo. Arrow indicates developing embryo residing in the gonad arm. Also indicated is a split egg with one half in the uterus developing (left) and the other half in the spermatheca that failed to develop (right). (H) Schematic of the C. elegans reproductive system.
nhr-6(lg6001) mutants are defective in spermatheca function and ovulation
Microscopic analysis of nhr-6(lg6001) homozygous adult hermaphodites revealed evidence of defective ovulation. In wild-type animals, ovulation involves a series of highly coordinated processes wherein the mature, most proximal oocyte in the gonad arm is moved into the spermatheca where it is fertilized (McCarter et al., 1999). After fertilization, the egg moves into the uterus where early embryonic development occurs prior to egg-laying (Fig. 2C). In nhr-6(lg6001) hermaphrodites, the proximal oocyte becomes fragmented during the ovulation process (Fig, 2D). Oocyte fragments were frequently observed in the proximal gonad arm (fragmented oocytes are not observed in regions distal to the most proximal oocyte), spermatheca, and uterus. Many fragments are fertilized and form an egg shell, indicating that the basis of the abnormal egg morphology phenotype is oocyte fragmentation and subsequent fertilization. 76% of nhr-6(lg6001) adult hermaphrodite gonads contained fragmented oocytes (n=25) when observed soon after mature oocytes begin to ovulate. Endomitotic oocytes are also observed in nhr-6(lg6001) gonad arms, further indicating that the ovulation process has been disrupted (Fig. 2E) (McCarter et al., 1999). Older nhr-6(lg6001) hermaphrodites display massive degeneration of gonad tissue (Fig. 2E). The basis for the degeneration is unknown. Tissue degeneration and the clogging of the gonad passages by oocyte fragments would ultimately prevent normal gonad function, including egg-laying, and would likely contribute to the bagging phenotype observed in nhr-6(lg6001) hermaphrodites.
Observations of ovulation in live nhr-6(lg6001) animals revealed that portions of ovulated oocytes often exit the spermatheca and re-enter the gonad arm. This process is likely to lead to oocyte fragmentation and subsequent initiation of endomitosis (McCarter et al., 1997). In addition, developing embryos are often observed within the gonad arm or spermatheca (Fig. 2E, G). 36% of nhr-6(lg6001) young adult hermaphrodites displayed this phenotype (n=25). The proportion of animals displaying misplaced developing embryos is increased in older hermpahrodites. Misplacement of developing embryos in the gonad arm or spermatheca and the re-entry of ovulated oocytes back into the gonad arm suggest that the proximal spermatheca valve and the distal spermatheca constriction do not function normally in nhr-6(lg6001) mutants. In fact, a spermatheca-uterine valve is not formed in nhr-6(lg6001) mutants (see below and Fig. 9). Both the proximal valve and distal constriction must open to allow passage of fertilized eggs and mature oocytes, respectively (see Fig. 5). The opening of these structures is tightly regulated and coordinated with the ovulation process (McCarter et al., 1999).
Figure 9. Defective spermatheca differentiation in nhr-6(lg6001) mutants.
In all micrographs distal is left and proximal is right. FITC-phalloidin of dissected adult gonads (A,B). (A) Dissected gonad from a wild-type adult. Note the circumferentially-oriented actin microfilaments in the spermathecal cells. The proximal oocyte of the gonad arm (thin arrow), the distal spermatheca constriction (#), the spermatheca sac containing a recently ovulated oocyte (thick arrow), and spermatheca-uterine valve (*) are indicated. (B) Dissected gonad from a nhr-6(lg6001) adult. Note the abnormal morphology and random pattern of microfilaments of the spermathecal cells. The distal constriction (#) does not have a constricted morphology. The spermatheca sac (thick arrow), also inflated with a recently ovulated oocyte appears to have missing cells, although it is not known if the cells were lost after ovulation or were missing prior to ovulation. Also note the absence of a spermatheca-uterine valve (*). (C, D) AJM-1::GFP expression in a young adult hermaphrodite bearing an integrated ajm-1::GFP transgene (jcIs1) (merged Nomarski and epifluorescence micrograph) in wild-type (C) and nhr- 6(lg6001) mutants (D). The bracket indicates the spermatheca region and the arrow the spermatheca-uterine valve. In both animals AJM-1::GFP illuminates the spermatheca lumen connecting the gonad arm and spermatheca-uterine border. Note decreased size of spermatheca and absence of spermatheca-uterine valve in nhr-6(lg6001) mutants. (E-H) MH27 (mouse monoclonal that recognizes AJM-1 protein) immunolocalization (E, F) and corresponding DAPI staining (G, H) of wild-type (E, G) and nhr-6(lg6001) (F, H) dissected young adult gonads. Arrows indicate distinct cells by DAPI stained nuclei. Note the cell size and shape differences of the cells in the proximal spermatheca of nhr- 6(lg6001), compared to wild-type. Arrowheads indicate diffuse, mislocalized AJM-1 protein in nhr-6(lg6001). Sp=sperm. In these micrographs, sperm is found in the lumen of the wild-type spermatheca but not the nhr-6(lg6001) spermatheca.
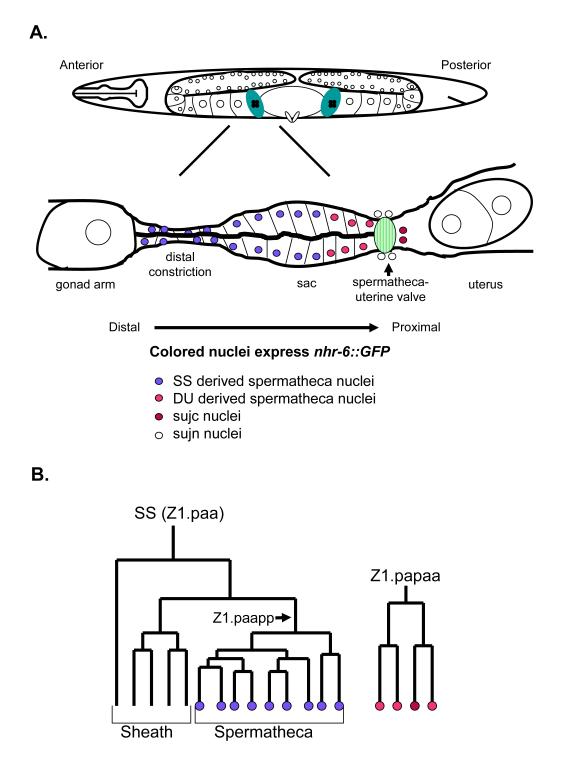
Figure 5. Schematic of the C. elegans spermatheca.
(A) The anterior spermatheca is shown for an adult hermaphrodite. Distal is left and proximal is right. The spermatheca is shown stretched out for clarity. Normally the spermatheca at this stage is a spiral structure (Kimble and Hirsh, 1979). The positions of nuclei are shown for diagrammatic purposes only and do not reflect the natural positions of spermatheca nuclei in adult hermaphrodites. Sperm are also not indicated for clarity. Colored nuclei express nhr- 6::GFP. Different colored nuclei represent different lineages contributing to the spermatheca. (B) One SS cell lineage and one DU lineage that give rise to the anterior spermatheca. Coloration corresponds to the nuclei shown in (A).
In addition to the absence of a spermatheca-uterine valve, the distal spermatheca constriction may also not function properly in nhr-6(lg6001) hermaphrodites. Ablation of cells in the distal constriction leads to fragmentation of oocytes (McCarter et al., 1997), similar to the fragmentation phenotype of nhr-6 mutants. Further evidence of abnormal distal spermatheca function was observed in feminized nhr-6(lg6001); fog-2(q71) adults. fog-2(q71) homozygous mutants are feminized and fail to generate sperm (Schedl and Kimble, 1988). Since ovulation is dependent on sperm signaling (Miller et al., 2001), oocytes in fog-2(q71) females are not ovulated or are ovulated at a decreased rate. The lack of ovulation in fog-2(q71) females leads to a compaction of oocytes in the ovary (Fig. 3A). fog-2(q71) adult females typically only display 1 or 2 unfertilized oocytes in the uterus. In contrast, all early nhr-6(lg6001); fog-2(q71) adult females contained a large mass of fragmented oocytes in the spermatheca and uterus (n=12; Fig. 3B). In addition, the compaction of oocytes within the gonad arm was not observed in these animals, indicating that oocytes were not contained in the gonad arm in the absence of sperm.
The nhr-6(lg6001); fog-2(q71) phenotype could be due to either a function for nhr-6 in the negative regulation of ovulation or to defective morphology of the distal constriction, which would also lead to a lack of oocyte containment. To assess spermatheca morphology, we performed nhr-6 RNAi in animals bearing an integrated fkh-6::GFP transgene (RNAi was necessary as the transgene is integrated into chromosome III, the same chromosome containing the nhr-6 locus). fkh-6 is a forkhead transcription factor that regulates sexual identity in C. elegans (Chang et al., 2004). fkh-6::GFP is expressed strongly in the developing and adult spermatheca (Chang et al., 2004). An RNAi sensitized rrf-3(pk1426);fkh-6::GFP was constructed and these animals were subjected to nhr-6 RNAi. Strongly affected animals (n=10) displayed severe defects in spermatheca morphology. These defects included decreased organ size, misplaced spermatheca nuclei, and a general disorganization that varied from animal to animal (Fig. 4B). Thus, it appears that abnormal spermatheca structure is a likely contributor to the defective ovulation and oocyte fragmentation phenotype. The expression of fkh-6::GFP indicates the spermatheca in nhr-6 loss of function animals is specified properly and that nhr-6 is not required for fkh-6::GFP expression.
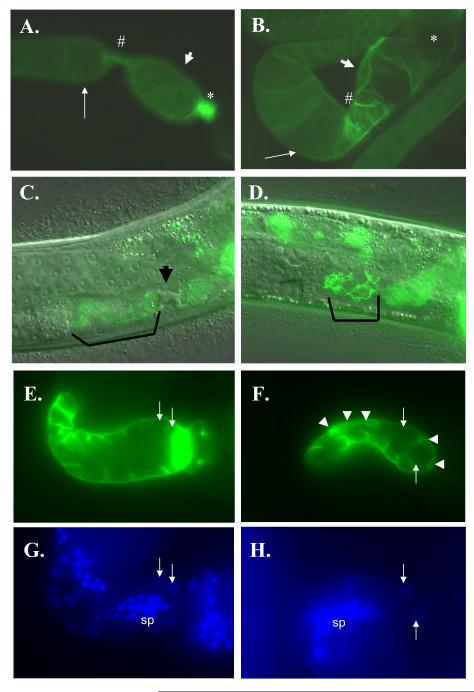
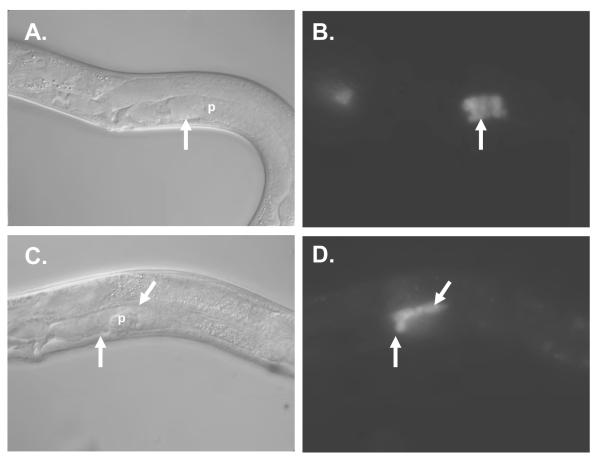
Figure 4. Expression of fkh-6::GFP in nhr-6(RNAi) hermaphrodites.
DIC (A, C) and epifluorescence (B, D) micrographs of fkh-6::GFP (A,B) and fkh-6::GFP; nhr-6(RNAi) (C, D) young adults. “p” indicates the proximal end of the gonad arm. Arrow indicates the spermatheca. The spermatheca in the fkh-6::GFP; nhr-6(RNAi) hermaphrodite is misshapen and spermatheca nuclei are observed over the dorsal side of the gonad arm, indicating loss of normal distal spermatheca organization.
Structure of the C. elegans spermatheca
The adult C. elegans spermatheca is composed of 24 epithelial cells (Lints and Hall, 2006). The apical surfaces of the epithelial cells face a lumen that expands as oocytes enter to become fertilized during ovulation. The lumen also stores mature sperm in gravid adults. The distal part of the spermatheca consists of eight cells that have a shape distinct from the cells that comprise the main part of the expandable sac (Fig. 5A). These specific cells make up a distal constriction that separates the gonad arm from the spermatheca. The cells of the distal constriction and sac contain a distinct pattern of circumferentially oriented actin filaments that allow the cells to expand during ovulation (McCarter et al., 1997). Another important feature of the C. elegans spermatheca is the spermatheca-uterine valve. At the young adult stage the valve constists of two syncytial structures. One structure is a 4-cell syncytial toroid that forms a tight, actin filament-rich valve separating the spermatheca from the uterus (Lints and Hall, 2006). Eggs fertilized in the spermatheca must pass through this valve structure to enter the uterus for further development. Another component of the valve is the spermatheca-uterine junction core. This syncytial structure contains two nuclei, the sujc, derived from the uterine lineages. In young adults, the sujc nuclei reside in the distal part of the uterus and extend filopodia through the spermatheca-uterine valve toroid into the spermatheca (Lints and Hall, 2006). The valve core is thought to be lost after the first egg passes from the spermatheca into the uterus. The precise function of the syncytial valve core is not known, although ablation of the sujc in L4 animals leads to reproductive defects (Palmer et al., 2002).
Each spermatheca is derived from two distinct cell lineages (Kimble and Hirsh, 1979). Eighteen of the 24 cells derive from the granddaughter cell of two SS cells. The SS cells are part of the early somatic gonad primordium and the daughters of these cells give rise to the gonad sheath and the distal and sac regions of the spermatheca (Kimble and Hirsh, 1979). The remaining six spermatheca cells are derived from the dorsal uterine (DU) lineage (Z1.papaa and Z4.apaaa for the anterior spermatheca; Z1.pappp and Z4. apapp for the posterior spermatheca). The sujc are also derived from these lineages (Fig. 5B). The sujn derive from a different dorsal uterine lineage.
nhr-6 is expressed in the developing somatic gonad and chemosensory neurons
To determine the expression pattern of nhr-6, we examined transgenic animals bearing a previously described nhr-6::GFP reporter construct (pCG52 (Gissendanner et al., 2004)) (Fig. 6). This transgene was chromosomally integrated and transgenic nematodes were analyzed at all developmental stages. As previously described for the extrachromosomal array containing this transgene (Gissendanner et al., 2004), we identified nhr-6::GFP expression in the integrated line in two cell types: a subset of somatic gonad cells in the L3 and L4 stages (Fig. 7) and a pair of head chemosensory neurons (likely to be ASI based on position) (Fig. 8). nhr-6::GFP was also weakly expressed in intestine in late embryos and L1 stage larvae (data not shown). Chemosensory neuron expression was first observed in three-fold embryos and was present in all larval stages and adults. The somatic gonad cells that express GFP in L4 animals include the developing spermatheca cells of the anterior and posterior spermathecae.
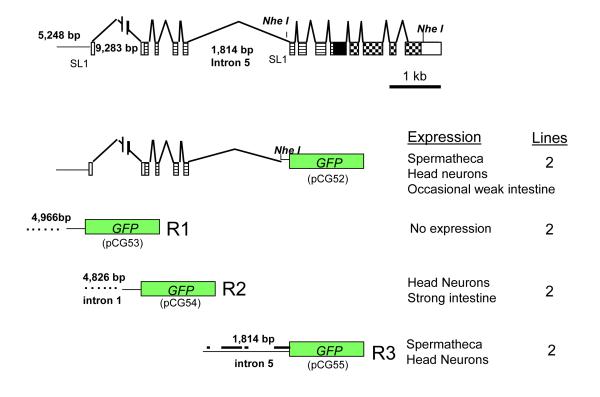
Figure 6. nhr-6::GFP reporter constructs.
nhr-6::GFP constructs and their correlation to the nhr-6 gene. pCG52 is considered to incorporate most if not all possible regulatory regions for nhr-6 as these regions were including in the nhr-6 genomic rescuing fragment. Transcriptional fusions (pCG53, 54, and 55) of the three potential regulatory regions are shown with their correlation to the full-length reporter. pCG53 contains 4,966 bp of sequence upstream of exon 1. pCG54 contains 4,826 bp of sequence (from intron 1) upstream of exon 2. pCG55 contains all of the sequence from intron 5. Blocks of sequence conservation between intron 5 of nhr-6 and the identical intron from the C. briggsae ortholog of nhr-6 is indicated by the thick bars in the pCG55 schematic. Only nhr-6 exon and intron sequences are drawn to scale. Sequences upstream of GFP in pCG53 and pCG54 are also not drawn to scale. The sizes in base pairs of these regions are indicated.
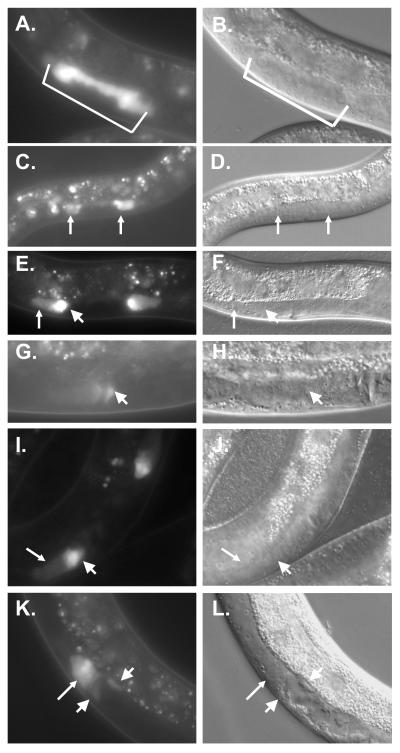
Figure 7. nhr-6::GFP expression pattern.
Animals in panels (A)-(H) and (K)-(L) bear an integrated full-length nhr-6::GFP transgene (nbIs1000). Corresponding DIC images are shown on the right for each epifluorescence micrograph. (A, B) Expression in 8 cells (bracket) of a late L3 nbIs1000 transgenic. (C, D) Expression in early L4 nbIs1000 transgenic. The arrows indicate the formation of anterior and posterior group of cells expressing the reporter. These cells are presumed to be spermatheca precursors. The animal in this picture is shown at a lower magnification than the other panels. (E, F) Mid-late L4 nbIs1000 transgenic nematode. Thin arrow indicates expression in the spermatheca and thick arrow indicates higher expression in the sujc cells. (G, H) Young adult nbIs1000 transgenic nematode. Arrow indicates diminishing expression in the sujc cells. (I, J) L4 nematode transgenic for the nhr-6R3::GFP reporter. Arrows indicate spermatheca and sujc cells as in (C). (K, L) Expression of nhr-6::GFP in nhr-6(lg6001) L4 hermaphrodite. Thin and thick arrows indicate the spermatheca and sujc as above. The sujc in the nhr-6(lg6001) mutants do not express brightly and are misplaced within the gonad.
Figure 8. Expression of nhr-6::GFP in anterior chemosensory neurons.
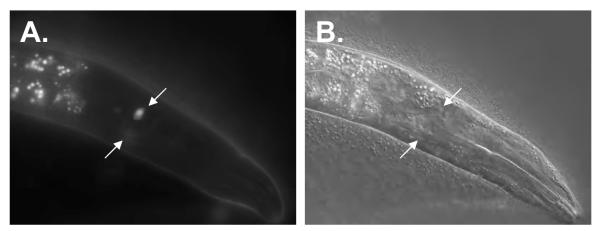
Epifluorescence (A) and DIC (B) micrographs of an nbIs1000 trangenic hermaphrodite. Expression in a pair of anterior chemosensory neurons is indicated by the arrows.
nhr-6 is expressed in the developing spermatheca and sujc
A more detailed analysis of developmental expression of nhr-6::GFP revealed initial somatic gonad expression in late L3 stage animals in 8 cells of the dorsal somatic gonad primordium (Fig. 7A, B). The positions are consistent with the cells being daughters of the DU and SS somatic gonad founder cells. We have not confirmed that the 8 initial cells that express nhr-6::GFP are the specific DU and SS daughter cells that give rise to the spermatheca. However, these GFP expressing cells increase in number and by early L4 they form two groups of cells that will later become the anterior and posterior spermathecae (Fig. 7C, D). The expression in the developing spermathecae continues throughout the L4 stage (Fig. 7E, F). Two cells in the proximal ends of each spermatheca display the brightest expression (Fig. 7E). These cells are the uterine lineage-derived sujc that form the core of the spermatheca-uterine valve (Kimble and Hirsh, 1979). nhr-6::GFP is not expressed in the sujn toroidal cells. During the L4/adult molt, expression in the spermatheca diminishes dramatically and in early adults only weak expression is observed in the differentiated sujc cells (Fig. 7G, H). No expression is observed in any spermatheca cells of gravid adults. The GFP expression patterns are also consistent with nhr-6 in situ hybridization data from the Y. Kohara laboratory (http://nematode.lab.nig.ac.jp/db2/ShowCloneInfo.php?clone=337g1).
Based on position and developmental stage, the cells that express nhr-6::GFP in the late L3 stage are likely to be SS and DU descendents that become spermatheca founder cells. Therefore, nhr-6::GFP appears to be expressed at the earliest stages of spermatheca development. The continued expression of nhr-6::GFP in the developing spermatheca, and the rapidly diminished expression of the reporter in late L4 suggests that nhr-6 has a developmental role in spermatheca function.
The function of nhr-6 in the somatic gonad appears to be specific to hermaphrodites. nhr-6::GFP expression was never observed in the developing and adult somatic gonad of male transgenics. Only head neuron expression was observed in males. Gonadal morphology of nhr-6(lg6001) mutant males was grossly normal and mutant males were capable of mating and generating cross progeny (see Materials and Methods).
The NHR-6B isoform is sufficient for spermatheca development
To further understand the regulation of nhr-6 expression, a series of transcriptional reporter fusions were constructed from three regions that could contain important regulatory elements. We have designated these putative regulatory regions R1, R2, and R3 corresponding to DNA sequence upstream of exon 1, exon 2 (intron 1) and exon 6 (intron 5), respectively (Fig. 6). Sequence from intron 1, immediately upstream of exon 2, was selected due to the large size of this intron, suggesting that it could contain regulatory elements. A reporter construct containing the first half of intron 1 was not constructed so we were unable to determine if regulatory elements reside in this part of the intron. We also reasoned that intron 5 could contain the regulatory elements necessary for expression of the nhr-6β isoform since intron 5 is fairly large (~1.8 kb) and lies immediately upstream of the SL1 splice site that defines the 5′ end of nhr-6β. We fused these potential regulatory regions to GFP and examined their expression patterns in transgenic animals. Expression of nhr-6R1::GFP was not observed in any cell type in two independent lines. nhr-6R2::GFP expression was observed in chemosensory neurons and the gut, but not in the developing or adult spermatheca. The nhr-6R3::GFP, however, was expressed in the developing spermatheca and sujc cells in a pattern identical to that of the original full-length reporter (Fig. 7I ,J). These data indicate that the important regulatory elements required for the spermatheca and sujc expression of nhr-6 reside in intron 5 of the gene. The alignment of nhr-6 sequence to the predicted nhr-6 ortholog in C. briggsae (C. briggsae predicted gene CBG09761), suggests that intron 5 of nhr-6 is conserved in C. briggsae (data not shown). Conserved blocks of sequence can be identified in this intron in sequence comparisons between nhr-6 and CBG09761 (Fig. 6).
Based on these results, we hypothesized that NHR-6B is the primary form expressed in the spermatheca. To further address this, we generated a rescue construct that only contains intron 5 and the downstream gene sequence of the shorter NHR-6B isoform. This construct fully rescued the reproductive phenotypes of nhr-6(lg6001) mutants (Table 1 and Table 2). This result demonstrates that the NHR-6B isoform, and the putative regulatory elements in intron 5 (and potentially in other downstream introns), are sufficient for normal spermatheca development. In addition, RNAi directed against the specific sequences of the longer nhr-6α mRNA isoform failed to generate any reproductive phenotypes (data not shown). Taken together, these data suggest a primary function for NHR-6B during spermatheca development but do not preclude expression or function of the larger isoform in the spermatheca.
We also tested auto-regulation of nhr-6 expression by crossing the integrated nhr-6::GFP into the nhr-6(lg6001) mutant background. We found that nhr-6::GFP seems to be expressed at normal levels in nhr-6(lg6001) mutants, suggesting that nhr-6 does not autoregulate in the developing spermatheca (Fig. 7K,L). However, bright sujc expression was not observed and the sujc were often found to be mislocalized in the gonad (Fig. 7K,L).
nhr-6 is required for proliferation of the spermatheca lineage
We made several observations of decreased spermatheca organ size (Fig. 4B, and Fig. 9D (below)) in nhr-6(lg6001) mutants. To determine if the decreased organ size was due to decreased number of cells, we performed several experiments to quantify spermatheca cell number in nhr-6 loss of function animals. In wild-type hermaphrodites at the young adult stage the spermatheca nuclei are easy to distinguish from other somatic gonad nuclei with DIC microscopy. However, since nhr-6(lg6001) hermaphrodites lack a spermatheca-uterine valve the border between the spermatheca and uterus is not easily distinguishable, limiting the usefulness of DIC analysis of young adult animals to count nuclei. Thus, alternative approaches were utilized. Spermatheca nuclei were first quantified using DAPI staining of extruded spermathcae. This analysis demonstrated that nhr-6(lg6001) spermathecae contain ~1/2 the normal number of spermatheca nuclei (Table 3). However, we found it difficult to obtain intact nhr-6(lg6001) spermathecae from the extrusion process, probably from the lack of organ integrity in the mutants. To further confirm the spermatheca cell number phenotype of nhr-6 (lg6001) we took advantage of the expression of nhr-6::GFP in nhr-6(lg6001) mutants. For this analysis, we used the nhr-6::GFP expression to outline to spermatheca organ in nhr-6(lg6001) in mid-late L4 hermaphrodites (spermatheca cell divisions are completed by this stage of development). Using optical sectioning, we then counted the spermatheca nuclei within the nhr-6::GFP expressing region (nhr-6::GFP has diffuse cellular expression). This analysis confirmed the decreased cell number phenotype of ~1/2 the normal number of spermatheca nuclei (Table 3). In addition, nuclei counts in young adults from the rrf-3(pk1426);fkh-6::GFP; nhr-6(RNAi) experiments further confirmed the cell number defect (Table 3).
Table 3.
Spermatheca nuclei counts1
| Experiment | N | # of nuclei | Range |
|---|---|---|---|
|
DAPI staining of extruded gonads N2 nhr-6(lg6001) |
6 6 |
24 ± 3 12 ± 1 |
20-27 11-14 |
|
nhr-6(+); nhr-6::GFP
nhr-6(lg6001); nhr-6::GFP nhr-6(lg6001); unc-26(e205)ced- 3(n717); nhr-6::GFP 2 |
21 35 10 |
24 ± 2 10 ± 1 10 ± 1 |
20-28 9-12 8-11 |
|
rrf-3(pk1426);fkh-6::GFP
rrf-3(pk1426);fkh-6::GFP; nhr-6(RNAi) |
5 10 |
24 12 ± 2 |
N/A 10-14 |
Spermatheca nuclei counts were performed by three methods: DAPI staining of extruded gonads, optical sectioning using nhr-6::GFP as a marker, and direct observation of nuclear localized fkh-6::GFP. There is some intrinsic error associated with the first two methods that lead to variability in the wild-type spermatheca nuclei counts (all wild-type spermathecae have 24 cells). In addition, the first two methods may sometimes include nuclei from the sujc or sujn. Regardless, nuclei counts for nhr-6(lg6001) were consistently decreased compared to wild-type. Unlike nhr-6::GFP, fkh-6::GFP is very tightly localized to spermatheca nuclei and is not expressed in the sujc or sujn; therefore, nuclei counts in wild-type background always revealed the normal 24 cell number.
unc-26(e205)ced-3(n717); nhr-6::GFP control animals were not directly counted for spermatheca nuclei but microscopic analysis of unc-26(e205)ced-3(n717) young adults revealed normal spermatheca size and morphology (data not shown).
The mammalian NR4A NR, Nur77, has been shown to have anti-apoptotic (pro-survival) properties (Suzuki et al., 2003). Therefore, one possibility is that the decreased cell number phenotype we observe could be due to apoptosis of spermatheca cells arising from the lack of nhr-6 activity in nhr-6(lg6001) mutants. Two observations argue against this possibility. First, we were unable to identify apoptotic (nor necrotic) cells in a detailed developmental analysis of spermathecae in nhr-6(lg6001) mutants (n=20, data not shown). Second, we constructed nhr-6(lg6001); ced-3(n717) double-mutants to genetically prevent apoptosis from occurring in nhr-6(lg6001) animals. nhr-6(lg6001); ced-3(n717) double-mutants were not suppressed for the reproductive defects (data not shown) nor the cell number defects (Table 3). Therefore, we conclude that nhr-6 is required for promoting cell proliferation of the spermatheca lineages, rather than preventing cell death, during spermatheca development.
nhr-6 is required for differentiation of the spermatheca
In nhr-6(lg6001) mutants the cells that form the spermatheca are specified but exhibit severe differentiation defects. In particular, nhr-6(lg6001) spermathecae have a disorganized cellular structure with a more random orientation of actin microfilaments, as indicated by staining with FITC labeled phalloidin (Fig. 9A,B). In addition, the distal constriction does not exhibit the constricted morphology seen in wild-type spermathecae (Fig. 9B).
To specifically assess epithelial differentiation in nhr-6(lg6001) spermathecae, we examined the expression of the AJM-1::GFP marker in wild-type and mutant nematodes. The ajm-1 gene encodes an adhesion junction molecule that is localized to adherens junctions of C. elegans epithelia, including the spermatheca (Aono et al., 2004; Koppen et al., 2001). In wild-type animals, the expression of AJM-1::GFP marks the apical, lateral, and basal cell junctions of spermatheca (Lints and Hall, 2006). Apical localization of AJM-1::GFP allows the visualization of the spermathecal lumen that connects the gonad arm to the spermatheca-uterine valve (Fig. 9C). In nhr-6(lg6001) mutants, the AJM-1::GFP marker is expressed, again demonstrating that spermathecal cells are specified in nhr-6(lg6001) mutants. A lumen can be observed in nhr-6(lg6001) mutants, as well as strong punctuate staining associated with AJM-1::GFP localization at the adherens junctions. The lumen of nhr-6(lg6001) spermatheca was smaller than wild-type, consistent with other observations of small spermathecae in nhr-6(lg6001) mutants (Fig. 9D). However, the expression of AJM-1::GFP was disorganized, and this disorganization was variable from animal to animal. The disorganization likely reflects cellular disorganization of the spermatheca in nhr-6(lg6001) mutants as cells of wild-type spermathecae have a highly ordered arrangement ((Lints and Hall, 2006) that is not observed in the mutant animals. Staining of extruded gonads with the monoclonal antibody MH27 (which recognizes the AJM-1 protein) revealed additional cellular defects in nhr-6(lg6001) mutants (Fig. 9F). These defects included cells that were either smaller or larger than wild-type spermathecae and a diffuse mislocalization of AJM-1 protein on the basal side of the cells. These phenotypes suggest specific differentiation defects in the spermathecal cells of nhr-6(lg6001) mutants.
The spermatheca and another somatic gonad structure, the gonadal sheath that surrounds the gonad, both derive from a common precursor cell, the SS cell. Two SS cells give rise to the gonadal sheath and the majority of the spermatheca cells of each gonad arm. Therefore, one possibility for the observed epithelial defects in nhr-6 (lg6001) mutants could be a partial misspecification (but not complete misspecification as spermatheca markers are expressed in the nhr-6(lg6001) hermaphrodites) of spermatheca cells to gonadal sheath cells. This does not appear to be the case as the spermatheca cells of nhr-6 (lg6001) animals fail to express myosin, a characteristic of the myoepithelial structure of the gonadal sheath (McCarter et al., 1997) (Fig. 10) as well as lim-7::GFP, a sheath-specific marker (Hubbard and Greenstein, 2000) (data not shown). Taken together, these data demonstrate a requirement for nhr-6 in the proper differentiation, but not specification, of the spermatheca organ. The morphology of the gonadal sheath, as well as the uterus, vulva, and ovary appear grossly normal in developing and young adult nhr-6(lg6001) mutants.
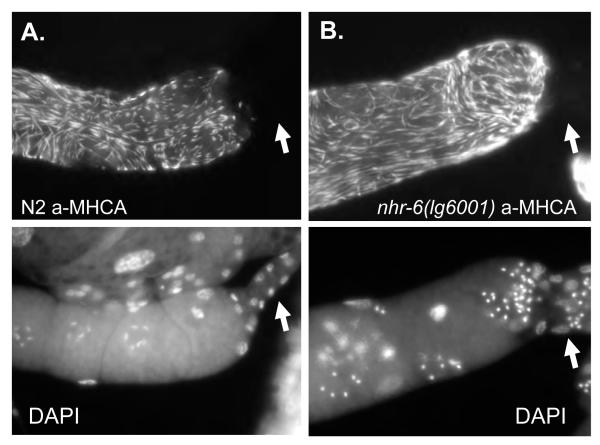
Figure 10. Expression of MHCA in wild-type and nhr-6(lg6001) gonads.
Epifluorescence micrographs of wild-type (A) and nhr-6(lg6001) (B) dissected early adult gonads stained with anti-MHCA (top) and DAPI (bottom). MHCA expression is restricted to the sheath of nhr-6(lg6001), indicating that nhr-6(lg6001) spermathecae have not adopted a sheath cell fate. Arrow indicates the spermatheca is present in the dissected gonad preparation. Note the lack of spermathecal distal constriction in the nhr-6(lg6001) gonad.
Another striking feature of the nhr-6(lg6001) phenotype is the completely penetrant absence of a spermatheca-uterine valve (Fig. 9C, D). Apparently, a connection between the spermatheca and the uterus can sometimes form in nhr-6(lg6001); however, this connection does not functionally separate the spermatheca and uterus as highly developed embryos are often observed in the spermatheca of nhr-6(lg6001) hermaphrodites (Fig. 2E).
The four cells that make up the valve derive from the uterine cell lineages and these cells have never been observed to express nhr-6::GFP. However, in young adults (prior to the first ovulation), these valve cells are closely associated with the sujc syncytium that forms the core of the valve. As noted above, nhr-6 is expressed in the sujc. A role for the sujc in the formation of the spermatheca-uterine valve has not been formally established. Nonetheless, one possibility for the observed absence of the spermatheca-uterine valve may be a cell nonautonomous role for nhr-6 in the development of the spermatheca-uterine valve.
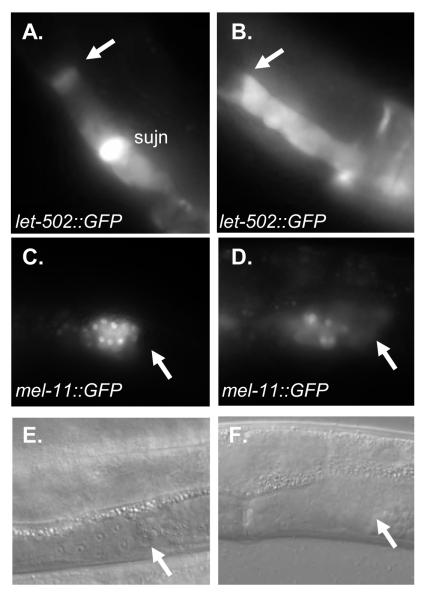
nhr-6 is required for the restricted spermathecal expression pattern of let-502
The epithelial and cytoskeletal defects associated with nhr-6 mutant spermathecae suggest that NHR-6 may be required for the normal expression of specific spermatheca differentiation genes. Very few genes have been described that have spermatheca development functions. Two of these genes are let-502 and mel-11. let-502 and mel-11 encode a Rho-associated kinase and myosin phosphatase regulatory subunit, respectively (Wissmann et al., 1999). These two proteins have counteracting functions in mediating rearrangements of the actin cytoskeleton. In the spermatheca, let-502 and mel-11 have non-overlapping domains of expression (Wissmann et al., 1999). let-502::GFP expression is restricted to the distal spermatheca and spermatheca-uterine valve while mel-11::GFP is specifically expressed in the spermathecal sac. To determine if the normal expression of these two genes requires nhr-6 function, we crossed the let-502::GFP and mel-11::GFP transgenic arrays (a generous gift from Paul Mains, University of Calgary) into the nhr-6(lg6001) background. We were unable to generate homozygous nhr-6(lg6001) animals bearing the let-502::GFP and mel-11::GFP transgenic arrays. Therefore, to observe expression of the transgene in animals lacking nhr-6 function we selected young adult transgenics segregated from nhr-6(lg6001)/+; Ex (extrachromosomal array bearing let-502::GFP or mel-11::GFP) animals. Homozygous nhr-6(lg6001) young adults were identified at high-magnification by screening for animals that lacked a spermatheca-uterine valve. Both markers were expressed in nhr-6(lg6001) mutants (Fig. 11). However, the let-502::GFP was misexpressed. Instead of expression being restricted to the distal and proximal regions of the spermatheca, let-502::GFP was expressed in all cells of the spermatheca organ in nhr-6(lg6001) young adults (n=7 spermathecae; Fig. 11A, B).
Figure 11. Expression of let-502::GFP and mel-11::GFP in nhr-6(lg6001) hermaphrodites.
Expression pattern of let-502::GFP (A, B) and mel-11::GFP (C, D) in wild-type young adult hermaphrodites (A, C) and nhr-6(lg6001) hermaphrodites (B, D). let-502::GFP expression is restricted to the proximal and distal regions in wild-type animals while expression is found in all spermathecal cells of nhr-6(lg6601) mutants. mel-11::GFP is expressed in the spermathecal sac of both wild-type and nhr-6(lg6001) mutants. Note the decreased number of expressing nuclei in nhr-6(lg6001) mutants. (E, F) DIC micrographs of animals in (C, D). Arrows indicate distal spermatheca. sujn: spermatheca-uterine valve. The spermatheca-uterine valve is not generated in nhr- 6(lg6001) mutants.
mel-11::GFP was normal in nhr-6(lg6001) mutants (restricted to the spermatheca sac). GFP expressed from the mel-11::GFP transgene is more tightly localized to spermathecal nuclei and, interestingly, and consistent with previous observations, the expression of mel-11::GFP was able to reveal a decreased number of spermathecal sac nuclei in nhr-6(lg6001) mutants. While the normal 16 spermathecal sac nuclei expressed the marker in wild-type mel-11::GFP transgenics (as determined by the presence of a normal spermatheca-uterine valve), only 8 + 1 GFP-expressing nuclei (range 7-10; n=6 spermathecae) were observed in nhr-6(lg6001) transgenics. In addition, one or more spermathecal nuclei distal to the expressing nuclei were consistently observed not to express GFP. Therefore, as in wild-type spermathecae, mel-11 also appears to have a proximally-directed expression in nhr-6(lg6001) mutants.
These data suggest that nhr-6 has a specific function in generating domains of let-502 gene expression important for spermatheca morphogenesis. This is potentially interesting given the apparent distal spermatheca defects of nhr-6(lg6001) spermathecae. Loss of proximal-distal polarity, as revealed by the let-502::GFP expression, could be a basis for distal defects. However, the alteration in let-502::GFP expression can also be explained by the cell proliferation phenotype of nhr-6(lg6001) spermathecae. As shown in Fig. 5, most of the spermatheca cells are derived from the SS lineage, including all of the cells of the distal constriction and the distal portion of the spermatheca sac. Therefore, the reduced number of distal cells in nhr-6(lg6001) spermatheca may contribute to the apparent loss of restriction in the expression of let-502::GFP.
nhr-6 is required for differentiation of the spermatheca-uterine junction core cells
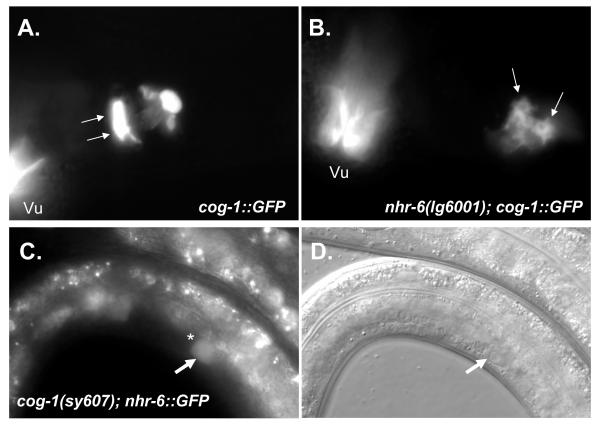
Given the absence of the spermatheca-uterine valve in nhr-6(lg6001) mutants, and the expression of nhr-6::GFP in the sujc cells that form the core of the valve, we assessed the formation and differentiation of the sujc cells in nhr-6(lg6001) animals using cog-1::GFP as a specific sujc marker. cog-1 encodes the C. elegans homolog of the Nkx6.1 homeobox transcription factor (Palmer et al., 2002). Like nhr-6, cog-1 mutants lack spermatheca-uterine valves (in addition to other gonad defects). We crossed an integrated cog-1::GFP reporter into the nhr-6(lg6001) background and assessed cog-1::GFP expression in 30 sets of sujc syncytia at the late L4 or young adult stage. Expression of cog-1::GFP was seen in 28/30 sets of sujc cells. 19 of these 28 displayed the normal number of 2 expressing sujc nuclei. Only one expressing sujc nucleus was seen in the remaining 9 sujc observations. cog-1::GFP expressing sujc cells were frequently misplaced in the somatic gonad and all sujc cells displayed differentiation defects. The level of cog-1::GFP expression was also frequently decreased. In young wild-type adults, the two nuclei of the sujc syncytium are found in the lumen of the uterus and filopodia extend across the valve lumen into the spermatheca (Fig. 12A). In nhr-6(lg6001) mutants, this normal morphology was never observed (Fig. 12B). Similar results were obtained with another sujc marker, lis-1::GFP (Dawe et al., 2001) (data not shown). Alterations in sujc formation and differentiation were never observed in nhr-6(+) hermaphrodites (n=30). These data indicate a function for nhr-6 in the differentiation, and possibly the specification, or generation, of the sujc syncytium.
Figure 12. Epistasis analysis nhr-6::GFP and cog-1::GFP.
(A,B) High-magnification epifluorescence micrographs of cog-1::GFP (syIs63) young adult hermaphrodite (A) and nhr-6(lg6001); cog-1::GFP young adult hermaphrodite (B). Arrows indicate the two sujc nuclei. Note in cog-1::GFP animals the filopodia that extend from the nuclei in the uterine lumen to the spermatheca. nhr-6(lg6001); cog-1::GFP hermaphrodites lack these filopodia and display an abnormal morphology, indicating abnormal differentiation of this structure in nhr-6(lg6001) mutants. Vu = cog-1::GFP expression in the vulva. (C, D) nhr-6::GFP (nbIs1000) expression in L4 cog-1(sy607) mutants. The star indicates loss of bright sujc expression (compare to Fig. 7E, F).
Since cog-1 also encodes a transcription factor, we were interested in whether nhr-6 expression requires cog-1. We crossed the integrated nhr-6::GFP transgene into the cog-1(sy607) mutant background. Interestingly, we did not observe nhr-6::GFP expression in the proximal region of the spermatheca (Fig. 12C,D). In wild-type animals, bright expression of nhr-6::GFP is observed in the proximal spermatheca in the cells that form the sujc (see Fig. 7E). Expression of nhr-6::GFP in the spermatheca proper was not affected. While this result supports an upstream role for cog-1 in the regulation of nhr-6::GFP expression, we were not able to confirm if the sujc are formed in cog-1(sy607) mutants. Thus, the loss of expression could be due to absence of the sujc as opposed to a specific loss of nhr-6::GFP expression. Intron 5 of nhr-6 does contain several TAAT-containing sequence elements that are similar to vertebrate Nkx6.1 binding sites (Jorgensen et al., 1999; Mirmira et al., 2000), and these could function as COG-1 homebox binding sites (data not shown).
DISCUSSION
NR4A is necessary for normal reproduction in C. elegans
We have demonstrated that the C. elegans NR4A nuclear receptor, nhr-6, is an essential gene for reproduction. Two mRNA and protein isoforms are expressed from the nhr-6 gene. The isoforms differ in the size of the N-terminal A/B domain and both would be expected to be able to bind DNA and activate transcription. Nematodes homozygous for a molecular null allele of nhr-6, lg6001, exhibit significantly reduced brood sizes and lay eggs that have abnormal morphology and frequently arrest development. The decreased brood size and abnormal egg morphology (and embryonic lethality) appear to arise from defective oocyte ovulation that results in the fragmentation of oocytes. This ovulation defect is a likely secondary consequence of abnormal spermatheca development. nhr-6 is expressed in the developing spermatheca and nhr-6(lg6001) mutants display an abnormal spermatheca morphology that would preclude normal ovulation. A striking feature of the nhr-6(lg6001) spermathecae is the absence of a proximal valve and morphologically abnormal distal constriction. The proximal valve and distal constriction have important roles in functionally separating the spermatheca from the uterus and ovary, respectively. This lack of separation likely prevents the ovulation process from being completed properly due to the ability of ovulated oocytes to re-enter the ovary. The lack of a spermatheca-uterine valve often leads to developing eggs residing in the spermatheca. Such situations would likely halt the reproductive process and lead to the “bagging” phenotype observed in mutant hermaphrodites.
We found that the shorter NHR-6 isoform, NHR-6B, is sufficient to direct normal spermatheca development. The N-terminal A/B domain has been shown to be important for the activity of NR4A NRs (Nordzell et al., 2004; Paulsen et al., 1992; Wansa et al., 2002). Since NR4A NRs appear to be true orphan receptors, post-translational modification of the A/B domain is likely to be critical in regulating NR4A activity. Rescue by the NHR-6B isoform identifies the C-terminal half the NHR-6 A/B domain as potentially important for NHR-6 activity. We also determined that the important regulatory elements driving nhr-6 expression in the spermatheca are found in the 1.8 kb intron 5, which lies directly upstream of the SL1 splice site that defines the beginning of nhr-6β. This intron appears to be conserved in C. briggsae and contains conserved blocks of sequence. This will facilitate the identification of upstream regulators of nhr-6.
Regulation of cell proliferation by NHR-6
In some contexts NR4A NRs have been shown to have mitogenic properties (Kolluri et al., 2003; Maxwell and Muscat, 2006; Nomiyama et al., 2006; Ponnio et al., 2002). Our data also suggest a role for NHR-6 in cell proliferation. Using a variety of approaches we have determined that the spermatheca of nhr-6 mutants have ~1/2 the normal number of cells. Our observations support the notion that the decrease in the number of cells is likely due to a failure in proliferation and not the activation cell death pathways. There are three possible explanations for the decrease in spermatheca cell number in nhr-6(lg6001) mutants. One explanation is not supported by our data: a cell fate transformation from spermatheca to sheath (both of which derive from a common precursor cell). Cell marker analysis demonstrates that the spermatheca is specified properly in nhr-6 mutants and, additionally, sheath cell markers are not ectopically expressed in mutant spermathecae. However, the cell proliferation defect could still be due to a lineage transformation. In each of the SS lineages that give rise to the spermatheca, the SS granddaughter cell that serves as a spermatheca founder cell undergoes an asymmetric division. As an example, the anterior spermatheca founder cell Z1.paapp divides to generate an anterior and posterior daughter (see Fig. 5B). The posterior daughter cell divides only once more to generate two spermatheca cells while the anterior daughter cell begins a more proliferative lineage that generates 7 distal spermatheca cells through three rounds of cell division. Thus, the decrease in cell number could be due to a transformation in the SS-derived spermatheca founder cell lineage (anterior→posterior for Z1.paapp). Such a transformation would decrease the number of distal spermatheca cells, leading to the formation of 11 total spermatheca cells (assuming all other divisions occur normally) and would explain the distal constriction morphology defects and the loss of polarity in let-502::GFP. The other possibility is that NHR-6 functions as a global regulator of cell proliferation and the observed cell number defect would thus be caused by a failure of cell divisions in all spermatheca lineages (both SS and DU derived). This latter possibility is better supported by the current data since global regulation would likely lead to animal to animal variability in cell number (which we observe) as well as a failure to produce cells from each lineage (which we also observe from the incompletely penetrant loss of sujc cells in nhr-6 (lg6001) mutants). Detailed lineage analysis will be required to determine which of these two models is correct.
nhr-6 may have multiple functions during spermatheca development
The epithelial defects of the spermathecae and the cell shape defects and disorganization of spermatheca and the spermatheca-uterine junction core (sujc) cells in nhr-6(lg6001) animals suggest strongly a differentiation function for nhr-6. Since nhr-6::GFP is expressed during the proliferative and differentiation stages of spermatheca development, it is tempting to speculate that nhr-6 may have dual functions in regulating cell proliferation and cell differentiation, possibly through the regulation of distinct gene sets in response to distinct cell-cell signals. A specific role for nhr-6 in cellular differentiation is supported by the sujc differentiation phenotype of nhr-6 mutants. The incompletely penetrant absence of the sujc in nhr-6(lg6001) animals indicates that nhr-6 functions in the generation of these cells. However, even when these cells are generated in nhr-6(lg6001) mutants they fail to display normal sujc morphology. Thus, NHR-6 likely functions in the transcriptional regulation of genes that direct sujc differentiation. A dual role for nhr-6 would be intriguing as it would position NHR-6 as a potential regulator of the cell proliferation to differentiation transition. The regulation of this transition in different organogenesis contexts is still very much an unsolved problem in developmental biology. The exact role of nhr-6 in spermatheca development will require careful temporal studies of gene function.
NHR-6 has a conserved function in organ development
The organogenesis role for nhr-6 is consistent with observed functions for NR4A nuclear receptors in vertebrates. NR4A NRs Nurr1 and NOR-1 have established roles in tissue differentiation and organ development, respectively, in mammals. All three NR4A NR paralogs have multiple biological roles in mammalian systems (Maxwell and Muscat, 2006). Despite this functional diversity, a common theme for the NR4A group is emerging. Each of the three mammalian paralogs (Nur77, Nurr1, NOR-1) regulate cell survival, cell proliferation, and/or cell differentiation in different biological processes (Castro et al., 2001; Kolluri et al., 2003; Martinez-Gonzalez et al., 2003; Ponnio et al., 2002; Ponnio and Conneely, 2004; Suzuki et al., 2003; Zeng et al., 2006; Zetterstrom et al., 1997). Nur77 has both pro-apoptotic and anti-apoptotic functions depending on the cellular context (Li et al., 2000; Suzuki et al., 2003). An ancestral function for this group may be the regulation of these cellular activities. The functions described here for nhr-6 are consistent with this notion. In C. elegans, the NHR-6 nuclear receptor appears to be regulating proliferation and differentiation during spermatheca development. Therefore, further studies of NHR-6 should enhance our general understanding of the cellular activities of this NR group.
The C. elegans spermatheca as a model system for organogenesis
Our study also illuminates the spermatheca as a new C. elegans model for the genetic regulation of organogenesis. The spermatheca has several unique features that argue for its utility as an organogenesis model. First, the spermatheca is a highly secretory organ and functions in signaling processes involving the gonad sheath and oocytes during ovulation (Bui and Sternberg, 2002; Clandinin et al., 1998). Signaling interactions with sperm are also evident as sperm are stored in the adult spermatheca and motile sperm from males preferentially migrate to this organ. In addition to this functional complexity, the spermatheca also exhibits structural complexity. The spermatheca is an endothelial-like tube with regulated valve connections to other tissues (ovary and uterus). This type of organ structure is commonplace in animals and genetic studies in C. elegans will provide insight into morphogenetic processes that shape this type of organ.
Our data also further establish the importance of the spermatheca-uterine valve core in organizing the C. elegans somatic gonad. It has been reported that the sujc are not generated in cog-1 mutants and a functional spermatheca-uterine valve fails to develop (Palmer et al., 2002). Likewise, a spermatheca-uterine valve is not generated in nhr-6(lg6001) mutants. Like cog-1, nhr-6 is expressed in the sujc but not in the uterine derived torroidal valve cells. Also, similar to cog-1 mutants, the sujc in nhr-6 mutants are either not generated or do not properly differentiate. Thus the transient sujc must have an important role in organizing the valve junction between the uterus and spermatheca, most likely through signaling interactions between the sujc and cells deriving from the uterine lineage. Our GFP reporter epistasis data suggest that NHR-6 functions downstream or in parallel to COG-1. As such, COG-1 is a good candidate to be an upstream regulator of NHR-6 during development of the sujc.
Acknowledgements
We thank David Greenstein, Paul Mains, Guy Caldwell, and Elegene AG (formerly of Martinsreid, Germany) for providing nematode strains; Yinhua Zhang from New England Biolabs for sharing an unpublished array integration protocol; and Brian Rowan for comments on the manuscript. Some strains were provided by the Caenorhabditis Genetics Center. This work was partly supported by start-up funds to C.R.G. from the Howard Hughes Undergraduate Education Program at ULM, grant P20 RR16456 under the INBRE Program of the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and grant LEQSF (2005-2008)-RD-A-39 to C.R.G. from the Louisiana Board of Regents. C.R.G. and C.V.M. gratefully acknowledge the support of Don Comb.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aono S, Legouis R, Hoose WA, Kemphues KJ. PAR-3 is required for epithelial cell polarity in the distal spermatheca of C. elegans. Development. 2004;131:2865–74. doi: 10.1242/dev.01146. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui YK, Sternberg PW. Caenorhabditis elegans inositol 5-phosphatase homolog negatively regulates inositol 1,4,5-triphosphate signaling in ovulation. Mol Biol Cell. 2002;13:1641–51. doi: 10.1091/mbc.02-01-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro DS, Hermanson E, Joseph B, Wallen A, Aarnisalo P, Heller A, Perlmann T. Induction of cell cycle arrest and morphological differentiation by Nurr1 and retinoids in dopamine MN9D cells. J Biol Chem. 2001;276:43277–84. doi: 10.1074/jbc.M107013200. [DOI] [PubMed] [Google Scholar]
- Chang W, Tilmann C, Thoemke K, Markussen FH, Mathies LD, Kimble J, Zarkower D. A forkhead protein controls sexual identity of the C. elegans male somatic gonad. Development. 2004;131:1425–36. doi: 10.1242/dev.01012. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, DeModena JA, Sternberg PW. Inositol trisphosphate mediates a RAS-independent response to LET-23 receptor tyrosine kinase activation in C. elegans. Cell. 1998;92:523–33. doi: 10.1016/s0092-8674(00)80945-9. [DOI] [PubMed] [Google Scholar]
- Dawe AL, Caldwell KA, Harris PM, Morris NR, Caldwell GA. Evolutionarily conserved nuclear migration genes required for early embryonic development in Caenorhabditis elegans. Dev Genes Evol. 2001;211:434–41. doi: 10.1007/s004270100176. [DOI] [PubMed] [Google Scholar]
- Giguere V. Orphan nuclear receptors: from gene to function. Endocr Rev. 1999;20:689–725. doi: 10.1210/edrv.20.5.0378. [DOI] [PubMed] [Google Scholar]
- Gissendanner CR, Crossgrove K, Kraus KA, Maina CV, Sluder AE. Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev Biol. 2004;266:399–416. doi: 10.1016/j.ydbio.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Hubbard EJ, Greenstein D. The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev Dyn. 2000;218:2–22. doi: 10.1002/(SICI)1097-0177(200005)218:1<2::AID-DVDY2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Jansen G, Hazendonk E, Thijssen KL, Plasterk RH. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat Genet. 1997;17:119–21. doi: 10.1038/ng0997-119. [DOI] [PubMed] [Google Scholar]
- Jorgensen MC, Vestergard Petersen H, Ericson J, Madsen OD, Serup P. Cloning and DNA-binding properties of the rat pancreatic beta-cell-specific factor Nkx6. FEBS Lett. 1999;461:287–94. doi: 10.1016/s0014-5793(99)01436-2. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–9. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Thummel CS. Nuclear receptors--a perspective from Drosophila. Nat Rev Genet. 2005;6:311–23. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- Kipreos ET. C. elegans cell cycles: invariance and stem cell divisions. Nat Rev Mol Cell Biol. 2005;6:766–76. doi: 10.1038/nrm1738. [DOI] [PubMed] [Google Scholar]
- Kolluri SK, Bruey-Sedano N, Cao X, Lin B, Lin F, Han YH, Dawson MI, Zhang XK. Mitogenic effect of orphan receptor TR3 and its regulation by MEKK1 in lung cancer cells. Mol Cell Biol. 2003;23:8651–67. doi: 10.1128/MCB.23.23.8651-8667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen M, Simske JS, Sims PA, Firestein BL, Hall DH, Radice AD, Rongo C, Hardin JD. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat Cell Biol. 2001;3:983–91. doi: 10.1038/ncb1101-983. [DOI] [PubMed] [Google Scholar]
- Li H, Kolluri SK, Gu J, Dawson MI, Cao X, Hobbs PD, Lin B, Chen G, Lu J, Lin F, Xie Z, Fontana JA, Reed JC, Zhang X. Cytochrome c release and apoptosis induced by mitochondrial targeting of nuclear orphan receptor TR3. Science. 2000;289:1159–64. doi: 10.1126/science.289.5482.1159. [DOI] [PubMed] [Google Scholar]
- Lints R, Hall DH. Reproductive system, part II. WormAtlas. 2006 http://www.wormatlas.org/handbook/reproductivesystem/reproductivesystem.htm.
- Maira M, Martens C, Batsche E, Gauthier Y, Drouin J. Dimer-specific potentiation of NGFI-B (Nur77) transcriptional activity by the protein kinase A pathway and AF-1-dependent coactivator recruitment. Mol Cell Biol. 2003;23:763–76. doi: 10.1128/MCB.23.3.763-776.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gonzalez J, Rius J, Castello A, Cases-Langhoff C, Badimon L. Neuron-derived orphan receptor-1 (NOR-1) modulates vascular smooth muscle cell proliferation. Circ Res. 2003;92:96–103. doi: 10.1161/01.es.0000050921.53008.47. [DOI] [PubMed] [Google Scholar]
- Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev Biol. 1997;181:121–43. doi: 10.1006/dbio.1996.8429. [DOI] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 1999;205:111–28. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. Vol. 48. Academic Press; San Diego, CA: 1995. pp. 451–482. [Google Scholar]
- Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291:2144–7. doi: 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- Mirmira RG, Watada H, German MS. Beta-cell differentiation factor Nkx6.1 contains distinct DNA binding interference and transcriptional repression domains. J Biol Chem. 2000;275:14743–51. doi: 10.1074/jbc.275.19.14743. [DOI] [PubMed] [Google Scholar]
- Newman AP, Acton GZ, Hartwieg E, Horvitz HR, Sternberg PW. The lin-11 LIM domain transcription factor is necessary for morphogenesis of C. elegans uterine cells. Development. 1999;126:5319–26. doi: 10.1242/dev.126.23.5319. [DOI] [PubMed] [Google Scholar]
- Newman AP, Inoue T, Wang M, Sternberg PW. The Caenorhabditis elegans heterochronic gene lin-29 coordinates the vulval-uterine-epidermal connections. Curr Biol. 2000;10:1479–88. doi: 10.1016/s0960-9822(00)00827-7. [DOI] [PubMed] [Google Scholar]
- Nomiyama T, Nakamachi T, Gizard F, Heywood EB, Jones KL, Ohkura N, Kawamori R, Conneely OM, Bruemmer D. The NR4A orphan nuclear receptor NOR1 is induced by platelet-derived growth factor and mediates vascular smooth muscle cell proliferation. J Biol Chem. 2006;281:33467–76. doi: 10.1074/jbc.M603436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordzell M, Aarnisalo P, Benoit G, Castro DS, Perlmann T. Defining an N-terminal activation domain of the orphan nuclear receptor Nurr1. Biochem Biophys Res Commun. 2004;313:205–11. doi: 10.1016/j.bbrc.2003.11.079. [DOI] [PubMed] [Google Scholar]
- Palmer RE, Inoue T, Sherwood DR, Jiang LI, Sternberg PW. Caenorhabditis elegans cog-1 locus encodes GTX/Nkx6.1 homeodomain proteins and regulates multiple aspects of reproductive system development. Dev Biol. 2002;252:202–13. doi: 10.1006/dbio.2002.0850. [DOI] [PubMed] [Google Scholar]
- Paulsen RE, Weaver CA, Fahrner TJ, Milbrandt J. Domains regulating transcriptional activity of the inducible orphan receptor NGFI-B. J Biol Chem. 1992;267:16491–6. [PubMed] [Google Scholar]
- Pekarsky Y, Hallas C, Palamarchuk A, Koval A, Bullrich F, Hirata Y, Bichi R, Letofsky J, Croce CM. Akt phosphorylates and regulates the orphan nuclear receptor Nur77. Proc Natl Acad Sci U S A. 2001;98:3690–4. doi: 10.1073/pnas.051003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnio T, Burton Q, Pereira FA, Wu DK, Conneely OM. The nuclear receptor Nor-1 is essential for proliferation of the semicircular canals of the mouse inner ear. Mol Cell Biol. 2002;22:935–45. doi: 10.1128/MCB.22.3.935-945.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnio T, Conneely OM. nor-1 Regulates Hippocampal Axon Guidance, Pyramidal Cell Survival, and Seizure Susceptibility. Mol Cell Biol. 2004;24:9070–8. doi: 10.1128/MCB.24.20.9070-9078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumar V, Chamberlin HM. The Pax2/5/8 gene egl-38 coordinates organogenesis of the C. elegansegg-laying system. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.068. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Maina CV, Gissendanner CR, Laudet V, Sluder A. Explosive lineage-specific expansion of the orphan nuclear receptor HNF4 in nematodes. J Mol Evol. 2005;60:577–86. doi: 10.1007/s00239-004-0175-8. [DOI] [PubMed] [Google Scholar]
- Rose KL, Winfrey VP, Hoffman LH, Hall DH, Furuta T, Greenstein D. The POU gene ceh-18 promotes gonadal sheath cell differentiation and function required for meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 1997;192:59–77. doi: 10.1006/dbio.1997.8728. [DOI] [PubMed] [Google Scholar]
- Schedl T, Kimble J. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics. 1988;119:43–61. doi: 10.1093/genetics/119.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears RC, Nevins JR. Signaling networks that link cell proliferation and cell fate. J Biol Chem. 2002;277:11617–20. doi: 10.1074/jbc.R100063200. [DOI] [PubMed] [Google Scholar]
- Sluder AE, Maina CV. Nuclear receptors in nematodes: themes and variations. Trends Genet. 2001;17:206–13. doi: 10.1016/s0168-9525(01)02242-9. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–56. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Suzuki N, Mirtsos C, Horacek T, Lye E, Noh SK, Ho A, Bouchard D, Mak TW, Yeh WC. Nur77 as a survival factor in tumor necrosis factor signaling. Proc Natl Acad Sci U S A. 2003;100:8276–80. doi: 10.1073/pnas.0932598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KD, Taylor LK, Haung L, Burlingame AL, Landreth GE. Transcription factor phosphorylation by pp90(rsk2). Identification of Fos kinase and NGFI-B kinase I as pp90(rsk2) J Biol Chem. 1999;274:3385–95. doi: 10.1074/jbc.274.6.3385. [DOI] [PubMed] [Google Scholar]
- Vidwans SJ, Su TT. Cycling through development in Drosophila and other metazoa. Nat Cell Biol. 2001;3:E35–9. doi: 10.1038/35050681. [DOI] [PubMed] [Google Scholar]
- Wang M, Sternberg PW. Pattern formation during C. elegans vulval induction. Curr Top Dev Biol. 2001;51:189–220. doi: 10.1016/s0070-2153(01)51006-6. [DOI] [PubMed] [Google Scholar]
- Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, Xu H, Walker NP, Perlmann T. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423:555–60. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- Wansa KD, Harris JM, Muscat GE. The activation function-1 domain of Nur77/NR4A1 mediates trans-activation, cell specificity, and coactivator recruitment. J Biol Chem. 2002;277:33001–11. doi: 10.1074/jbc.M203572200. [DOI] [PubMed] [Google Scholar]
- Wingate AD, Campbell DG, Peggie M, Arthur JS. Nur77 is phosphorylated in cells by RSK in response to mitogenic stimulation. Biochem J. 2006;393:715–24. doi: 10.1042/BJ20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissmann A, Ingles J, Mains PE. The Caenorhabditis elegans mel-11 myosin phosphatase regulatory subunit affects tissue contraction in the somatic gonad and the embryonic epidermis and genetically interacts with the Rac signaling pathway. Dev Biol. 1999;209:111–27. doi: 10.1006/dbio.1999.9242. [DOI] [PubMed] [Google Scholar]
- Zeng H, Qin L, Zhao D, Tan X, Manseau EJ, Van Hoang M, Senger DR, Brown LF, Nagy JA, Dvorak HF. Orphan nuclear receptor TR3/Nur77 regulates VEGF-A-induced angiogenesis through its transcriptional activity. J Exp Med. 2006;203:719–29. doi: 10.1084/jem.20051523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterstrom RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276:248–50. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]