Abstract
The Human Connectome Project (HCP) has developed protocols, standard operating and quality control procedures, and a suite of informatics tools to enable high throughput data collection, data sharing, automated data processing and analysis, and data mining and visualization. Quality control procedures include methods to maintain data collection consistency over time, to measure head motion, and to establish quantitative modality-specific overall quality assessments. Database services developed as customizations of the XNAT imaging informatics platform support both internal daily operations and open access data sharing. The Connectome Workbench visualization environment enables user interaction with HCP data and is increasingly integrated with the HCP's database services. Here we describe the current state of these procedures and tools and their application in the ongoing HCP study.
Introduction
The Human Connectome Project (HCP) is midway through an unprecedented 5-year effort to obtain high resolution structural, functional (fMRI), and diffusion (dMRI) imaging data from each of 1200 subjects. After an initial 2-year period of refining the methods for data acquisition and analysis, a 3-year effort to systematically acquire, process, and share these data broadly with the research community begin in August, 2010. In this article, we describe several informatics-related aspects of the project that are pivotal to its overall success: quality control (QC) procedures, database services, and data visualization.
The QC procedures used by the HCP build on approaches developed by previous large scale imaging projects such as the Biomedical Informatics Research Network (BIRN) (Helmer et al., 2011) and Alzheimer's Disease Neuroimaging Initiative (ADNI) (Jack et al., 2008). These include the use of documented standard operating procedures, pre-acquisition scanner monitoring, post-acquisition manual and automated image review, and automated verification of scan acquisition parameters. Here, we describe the implementation of these QC procedures in the HCP and demonstrate their utility in executing a high throughput large scale imaging study.
The database services developed by the HCP focus on supporting the program's daily operations, including the QC procedures described above, plus its commitment to user-friendly data sharing. The HCP methods introduce unique challenges unlike those encountered in previous large-scale neuroimaging projects. The acquisition protocol itself yields over 200 minutes of high resolution data from each participant, including nearly 9000 frames (image volumes) of 2 mm resolution fMRI data and 576 volumes of 1.25 mm resolution dMRI data per subject. The processing streams in development by the HCP produce multiple dense connectomes, each of which exceeds 30 GB in size. In total, the HCP storage requirements are expected to approach 1 petabyte, or the equivalent of about 220,000 DVDs. The database services being implemented by the HCP are largely aimed at managing the storage, processing, and distribution of these massive volumes of data. In addition, we are developing novel informatics capabilities to enable advanced data exploration, mining, and visualization. Here we describe the work on these fronts that has been completed to date and provide a preview of additional developments that are underway.
Overview
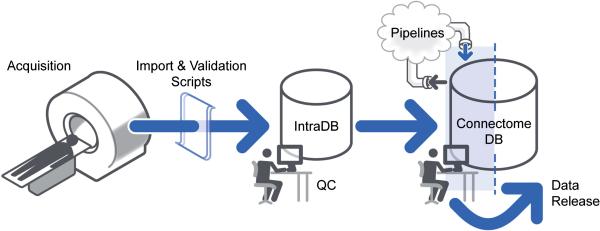
The overall HCP workflow from data acquisition to data sharing is managed by a comprehensive informatics infrastructure schematized in Figure 1. An internal private database system (IntraDB) - an XNAT-based system (Marcus et al., 2007) that has been customized to support the HCP's data types and workflows - manages the HCP's daily operations and quality control procedures. For each of the HCP's different data acquisition systems, a custom import script transforms the acquired data from system-specific formats to XNAT-compliant XML and posts the generated XMLs to IntraDB through XNAT's web service programming interface (API). The manual and automated QC procedures described below are all conducted within IntraDB. Automated QC procedures are implemented using the XNAT pipeline service. Once data have passed QC, they are transferred to ConnectomeDB, a second XNAT-based system that has been highly customized for the HCP's data sharing and data mining services. Data in ConnectomeDB are organized into projects that align with the HCP's quarterly data release schedule. Prior to public release the project for a given quarter are accessible only to internal HCP staff for review and execution of the HCP's image processing pipelines (Glasser et al., this issue). Once the data have been released, the project is made accessible to all registered ConnectomeDB users. For more details on the HCP's data sharing approach and policies, see Van Essen et al (this issue).
Figure 1. The HCP data flow.
HCP data are acquired on a dedicated scanner and immediately sent to IntraDB, where a series of manual and automated quality and anonymization processes are executed. Data that pass quality control are re-organized and exported to ConnectomeDB for public sharing.
Quality Control
The success of the project hinges fundamentally on the quality and consistency of the collected data. To this end, the HCP has implemented multiple levels of quality control, ranging from real-time oversight during acquisition to post-acquisition manual and automated image review. Each of these procedures is specified in a formal standard operating procedure and integrated into the internal database system.
Standard operating procedures (SOPs)
An important aspect of our quality planning has been the creation of Standard Operating Procedures (SOPs). HCP SOPs are step-by-step, written instructions on how to perform each aspect of HCP data acquisition. Our current SOPs cover the procedures to be followed when participants first arrive, the behavioral testing sessions and each of the 3T MRI imaging sessions. Future SOPs will also cover MRI scans at 7T and MEG/EEG scans to be carried out on a subset of HCP participants.
The primary purpose of the SOPs is to ensure consistency in data acquisition over the course of the study, since HCP's 1200 subjects are being scanned over a period of 3 years. SOPs are scheduled for quarterly reviews by research staff to ensure that procedures do not ‘drift’ over time. They also serve as a training aid when new research staff members are hired, and as a basis for internal HCP auditors’ assessments as to whether research staff members are administering tests in a consistent manner. In addition to their important role in promoting consistency, HCP's SOPs have also been posted at www.humanconnectome.org, where they serve as documentation for the scientific community on how our data are being collected.
Scanner maintenance
It is important that the customized Connectome Skyra scanner provide stable performance over the 3-year course of the main HCP acquisitions. To ensure that the scanner remains properly calibrated throughout the study, a Siemens engineer familiar with the customizations of the Connectome Skyra conducts a preventative maintenance visit every 3 months to ensure that the scanner remains “within specification”. Also, on a frequent (near daily) basis, the local MR technologist runs the “Head 32 Coil Check” routine of Siemen's quality assurance (QA) procedures to verify that the signal-to-noise of each coil element remains within specification. Notably, problems with individual coil elements do not necessarily manifest visibly in reconstructed structural, fMRI, or dMRI images; on multiple occasions this coil check has already revealed problems with individual coil elements that were promptly fixed. On a similarly frequent (near daily) basis, the MR technologist also scans an Agar phantom, collecting 400 time points (4.8 min) of data using the same parameters employed for the human fMRI scans (i.e., TR=720 ms, TE=33.1 ms, flip angle=52°, 2 mm isotropic voxels, multiband acceleration factor of 8). These data are used to assess scanner stability and are automatically processed within the IntraDB using the BIRN Agar Phantom QA tool (fmriqa_phantomqa.pl1, using a 30x30 voxel ROI in the center of the middle slice of the volume) (Friedman and Glover, 2006). The ensuing HTML report (including various QA-related plots and images) is viewable within IntraDB for internal review and will be made available for open access review in ConnectomeDB. The outputs of this BIRN tool include quantitative metrics of temporal SNR (tSNR), static image SNR, percent signal fluctuation and drift, scanner stability (Weisskoff, 1996), smoothness, and percent intensity of ghosting, which are aggregated for tracking across time. Figure 2 shows the tSNR, SNR, and ghosting intensity values from the Agar scans over a 7-month period.
Figure 2. Selected measures from the BIRN Agar phantom QA over a 7-month period.
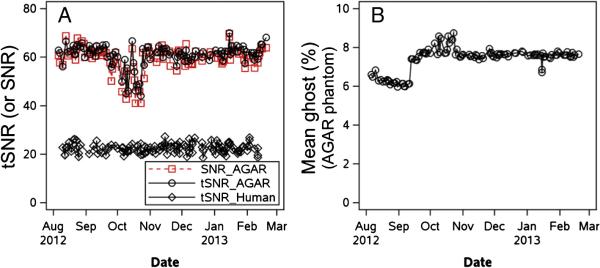
Panel (A) shows temporal SNR [tSNR; also called the summary signal-to-fluctuation-noise value in Friedman and Glover (2006)] and static image SNR in the Agar phantom. tSNR from one resting-state scan of each human participant over this time window is also shown. Panel (B) shows the percent ghost intensity, calculated by shifting an original phantom mask by N/2 voxels in the appropriate axis to create a “ghost mask” and then computing the mean intensity in that mask relative to the non-ghost voxels.
tSNR and SNR in the phantom track each other closely, indicating excellent within-run scanner stability (i.e., tSNR is determined by the intrinsic static image SNR). There was a period of increased variability in tSNR and SNR in the phantom in October 2012 (Figure 2a), which the Siemens engineer suspected was caused by variations in room humidity as the heating/cooling systems fluctuated in the fall weather of St. Louis. However, tSNR in the human fMRI scans (see below) remained steady over this period (lower line in Figure 2a), indicating that the effect seen in the Agar phantom scans did not affect the quality of the human fMRI acquisitions. Figure 2b shows that the ghosting level of the Agar phantom scans increased slightly in September 2012, which was the consequence of a hardware board replacement required to implement a gradient smoke detector safety feature. These plots illustrate the value of the Agar phantom QC in monitoring and ensuring stable scanner performance over time.
Data acquisition
The design of the HCP study population and the inclusion and exclusion criteria for prospective participants are described elsewhere (Van Essen et al., 2013, this issue). After three members of a family have passed screening, each participant is assigned a 6-digit identifier in which the first four digits are a randomly generated number and the last two digits are a checksum of the first four digits. This scheme ensures that errant entry of any single digit of the subject ID in any of the HCP data systems would result in an invalid ID rather than a match with another subject ID. The subject ID is automatically imported from the recruitment data system into IntraDB on a nightly basis.
Participants come to Washington University for the full HCP testing protocol, typically obtained over a consecutive two-day visit. Behavioral data are collected using the web-based NIH Toolbox Assessment Center (http://www.nihtoolbox.org) and a modified web-based Penn computerized battery (Gur et al., 2001; Gur et al., 2010). The data generated by these systems are automatically imported into IntraDB.
A number of HCP's data acquisition QA measures focus on ensuring that participant head motion is minimized during MR scans. Prior to the initial acquisition, participants are positioned in a mock scanner to get acclimated to the scanner bore. While in the mock scanner (for approximately 10-15 minutes), participants are trained to keep their head still using motion tracking software. During the actual scan, the head is stabilized using cushions on the sides and top of the head. During structural and diffusion sessions, participants watch a movie. The movie pauses for 5 seconds when head motion exceeds a specified threshold, providing immediate feedback to participants that they have moved too much. To the extent permitted by the computational demands of multiband image reconstruction, the technologist visually inspects all images as reconstruction completes and immediately reacquires the scan if it is not deemed to be of sufficient quality.
Data are transferred to IntraDB at the completion of each imaging session and image reconstruction process using the standard DICOM transfer protocol. Once the data are received by IntraDB, several processes are triggered, including automated acquisition parameter checks, image “defacing,” file format conversion from DICOM to NIfTI, email notifications, and automated quantitative quality control pipelines. Image defacing (obscuring of identifying features, including the ears) is executed on all high resolution structural sequences (Milchenko and Marcus, 2013). The study coordinator conducts a daily review of these processes to identify missing data, resolve discrepancies, and schedule additional imaging sessions with the study participants as needed.
Preservation of unreconstructed (‘raw’) image data
Reconstruction of raw multiband image data is a complex process that undergoes ongoing algorithmic improvements (Ugurbil et al., 2013, this issue?). The HCP will strive to balance the benefits of reconstruction improvements against the desirability of reconstruction consistency across subjects. Following the commencement of HCP Phase II scanning in August 2012, an improved reconstruction algorithm was developed that reduced spatial blurring in fMRI and dMRI acquisitions. This improvement was deemed worthy of upgrading the reconstruction algorithm, which occurred in April 2013. The new reconstruction method will be applied retrospectively to existing dMRI scans that were saved in a compact but unreconstructed data format. (These unreconstructed volumes are available for the vast majority of dMRI sessions before the transition). For fMRI scans prior to April 2013, unreconstructed scans were not preserved owing to technical considerations related to the time required to complete reconstruction at the scanner. However, concurrent with the change in reconstruction algorithm in April 2013, the reconstruction switched to using graphics-processing-unit (GPU) accelerated reconstruction, which reduces reconstruction times considerably, thus permitting saving of unreconstructed fMRI data from April 2013 onward. Details regarding which reconstruction was used will be clearly communicated as part of HCP quarterly releases. Additionally, for fMRI (for which retrospective reconstruction of scans prior to April 2013 is not available) the HCP will provide datasets processed using both reconstruction algorithms for a small subset of subjects.
Parameter extraction and acquisition validation
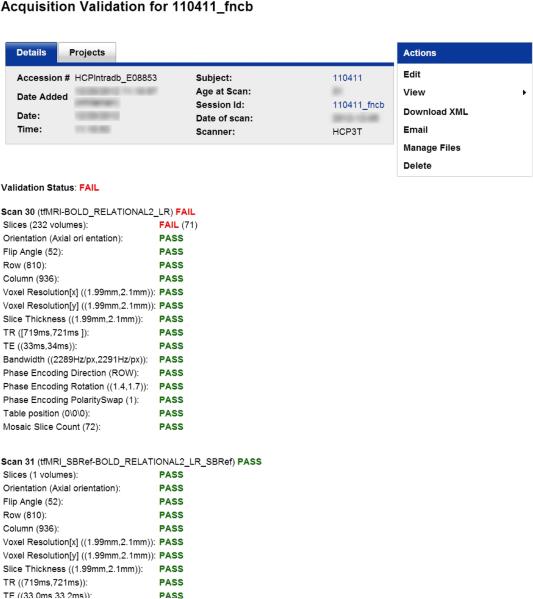
Numerous DICOM fields are extracted and checked against their expected value as part of a Protocol Validation pipeline, immediately following transfer of the DICOMs from the Skyra to the IntraDB. These checks include subject position (i.e., “head-first-supine”), table position, voxel resolution, image dimensions, acquisition orientation, TR, TE, TI, flip angles, bandwidth, phase encoding direction and polarity, number of DICOMs received, and diffusion b-value maximum, as appropriate for each scan type. The pipeline posts a detailed report to IntraDB (Figure 3). Any parameter mismatches are clearly flagged and an automatic email is sent to HCP staff for investigation and follow-up. This process provides protection against any inadvertent protocol changes at the scanner (since there is no convenient mechanism for “locking” a USER-tree protocol), as well as data corruption introduced during data transfer, anonymization, and data storage. In practice, no inadvertent protocol changes or data corruption have been observed to date, and the Protocol Validation pipeline therefore primarily serves to flag scans which were aborted at the scanner or which incurred an error during the image reconstruction process. The scanner operators review this immediate Validation pipeline when a parameter mismatch is reported. When images are missing from a scan, the scanner operators either indicate in IntraDB that such images are permanently missing (e.g., in the case of scanner acquisition or image reconstruction problems), or repeat the reconstruction and re-upload the missing images if possible. Additionally, other parameters are also extracted from the DICOMs into the database for informational purposes and possible use in subsequent processing, including echo spacing, sequence versioning information, a UUID linking each dMRI and fMRI scan to its associated physiological (heart rate and respiration) data, and the linear and quadratic shim currents in effect at the time of the scan so that the equivalence of the shim field between scans can be established.2
Figure 3. Protocol validation reports.
Reports are generated immediately after acquisition to detail whether each series with the session was acquired according to the HCP protocol. Here, the series 30 scan failed owing to an insufficient number of frames, likely due to scan being aborted prior to completion.
QC of structural MRI
Since all other modalities are dependent on high quality structural data, the HCP protocol was designed to schedule the structural imaging session first, allowing the data to be quickly reviewed and reacquired if the initial scans are inadequate in quality. Technicians review image quality during the scan session. Most issues are caught immediately, and problematic scans can often be reacquired within the same session. Subsequently, QC review personnel are automatically notified immediately after the structural session is transferred to IntraDB. They perform a multi-point review process, within hours of acquisition when feasible. The review includes inspection for image crispness, blurriness, motion and other artifacts, and accuracy of defacing. Each T1w or T2w scan is assigned an overall numerical rating on a 4 point scale (1=poor, 2=fair, 3=good, 4=excellent). If the reviewed session does not include at least one T1w scan and at least one T2w scan with ratings of “good” or higher, the reviewer informs the study coordinator, who schedules an additional imaging session for the participant, which is typically obtained on the participant's Day 2 visit. Combining structural scans across sessions is avoided because it is important that they be linked to the same bias field and B1 field map during preprocessing. Weekend scans are formally reviewed on the following Monday, though initial rescan determinations can also be made by the technologists running the scanner at the time of acquisition.
QC of fMRI
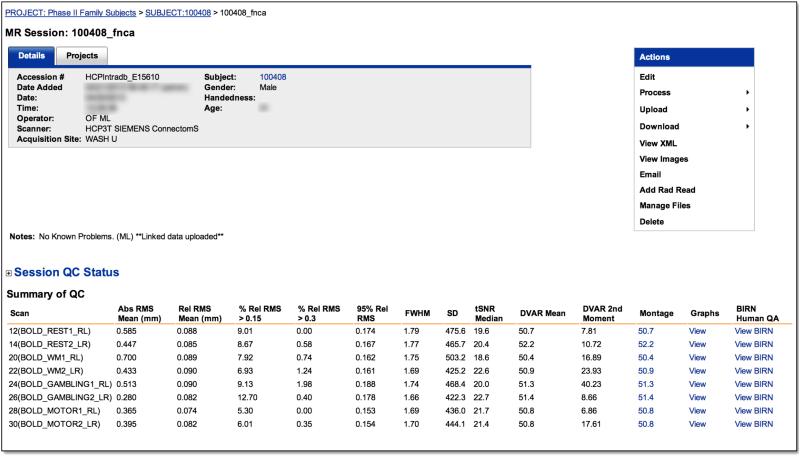
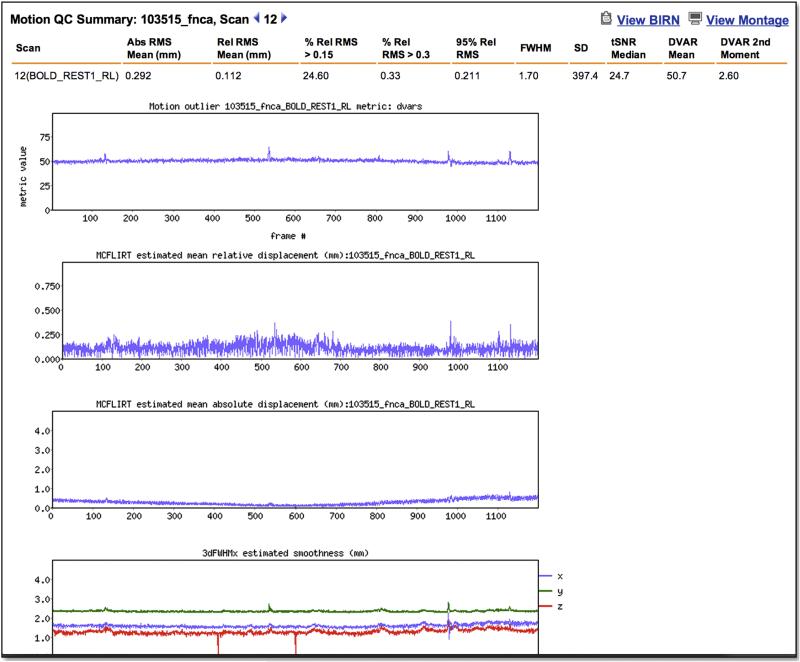
A “FUNCTIONAL_QC” pipeline was implemented to (i) perform rapid quality control (QC) to identify scans that are potential candidates for re-acquisition (i.e., before the participant leaves on Day 2), (ii) facilitate retrospective review of large numbers of fMRI scans within the IntraDB, and (iii) eventually support queries and subject filtering within the public ConnectomeDB based on scan quality. Immediately after conversion of the raw DICOMs to NIFTI format, the FUNCTIONAL_QC pipeline is launched for QC of both task and resting-state fMRI scans. Note that because this pipeline is intended for rapid QC and feedback, it is appreciably simpler than the full “fMRIVolume” pipeline (Glasser et al, this issue) that generates the “minimally preprocessed” fMRI data distributed by the HCP. The FUNCTIONAL_QC pipeline starts with motion correction (using spline interpolation) to the central time point via FSL's ‘MCFLIRT’, brain extraction via ‘BET’, and intensity normalization of the entire 4D time series dataset by a single multiplicative factor (to a grand median of 10000). The voxel-wise temporal standard deviation (tSD) and temporal SNR (tSNR; mean over time divided by standard deviation over time) (Triantafyllou et al., 2005) are computed, yielding tSD and tSNR images. Smoothness (FWHM, in mm) is computed for each frame in the x, y, and z directions, as well as their geometric mean, using AFNI's ‘3dFWHMx’. Finally DVARS (Power et al., 2012), the root mean squared change in BOLD signal between succcessive image volumes, is computed for each frame using FSL's ‘fsl_motion_outliers’. Outputs of this pipeline include the images of tSNR and tSD, and plots of movement rotation and translation, absolute movement (relative to the central reference frame), relative movement (relative to the preceding frame), smoothness, and DVARS over the frames (time points) of the run, all of which are easily viewable within the IntraDB accessible to HCP staff (Figure 4). Various quantitative summary metrics are also computed for each run and displayed in tabular format in the IntraDB, including mean of the absolute and relative movement time series; percentage of frames with relative movement above various thresholds; 90th and 95th percentile values of the relative movement time series; mean and standard deviation of the DVARS time series; and the median tSD and tSNR within the brain mask. Additionally, each run is processed through the BIRN Human QA tool (Glover et al., 2012) and the HTML report it generates is viewable within the IntraDB as well.
Figure 4. Quality control reports.
IntraDB includes detailed reports for reviewing data and quality control information. A session-level report (top) provides modality-specific summary statistics for each scan. For fMRI, this includes the output of the FUNCTIONAL_QC pipeline (temporal SNR, DVARS, FWHW, etc). Detailed plots of these measures across time (bottom) can be viewed by clicking on a link within the report. The full report generated by the BIRN QA pipeline can also be accessed.
Currently we are establishing normative values for these fMRI QC metrics in the context of the HCP fMRI acquisitions. Table 1 shows statistics for nine of the measures based on 2207 fMRI runs (from 126 subjects). To achieve full brain coverage with increased spatial and temporal resolution the HCP acquisition protocol makes use of multiband sequences (Ugurbil et al., this issue). Due to the smaller voxels (2 mm isotropic) and shorter TR (720 ms) of the HCP fMRI acquisitions compared to most other studies, the tSNR averaged over the whole brain of the HCP acquisition is smaller than has been reported in other studies3 (Triantafyllou et al., 2005, Greve et al., 2011, Hutton et al., 2011). Our acquisitions operate in a regime of higher thermal and system noise relative to most other studies – a regime where thermal and “physiological” noise are predicted to be roughly equal in gray matter (Bodurka et al., 2007, Triantafyllou et al., 2011). The increased spatial resolution of the HCP protocol supports more accurate localization of the fMRI data to the cortical surface (Glasser et al., this issue), and the increased temporal resolution supports improved methods of denoising (Smith et al., this issue) and novel analyses of brain activity (Smith et al., 2012). Importantly, while the tSNR of the HCP acquisitions is lower than other studies with larger voxels and longer TRs (both of which are associated with increased tSNR), we still achieve high quality mapping of task and resting-state activity within individual subjects (Barch et al., Smith et al., this issue).
Table 1.
Distribution of Functional QC measures.
| Measure | Meana | Std Dev | 5th percentile | 95th percentile |
|---|---|---|---|---|
| tSNRb | 24.41 | 2.39 | 20.37 | 28.04 |
| FWHM: mean (mm) | 1.66 | 0.080 | 1.56 | 1.82 |
| AbsMotion: mean (mm) | 0.31 | 0.21 | 0.13 | 0.72 |
| RelMotion: mean (mm)c | 0.093 | 0.029 | 0.06 | 0.15 |
| RelMotion: 95th percentile (mm) | 0.19 | 0.070 | 0.12 | 0.30 |
| RelMotion > 0.15 (%)d | 15.55 | 13.97 | 1.14 | 44.69 |
| RelMotion > 0.30 (%)e | 1.00 | 2.42 | 0.00 | 5.20 |
| DVARS: mean (Δ% BOLD × 10)f | 52.86 | 3.95 | 47.17 | 59.93 |
| DVARS: std dev | 2.98 | 2.50 | 1.01 | 6.85 |
N = 2207 task and resting-state fMRI runs (126 subjects)
temporal signal-to-noise ratio (mean divided by temporal standard deviation), computed voxelwise, then averaged over a whole brain mask
Relative motion as returned by FSL's MCFLIRT is approximately 1/2 of the “Framewise displacement” (FD) measure defined in Power et al. (2012).
Percentage of frames in the time series with relative motion > 0.15
Percentage of frames in the time series with relative motion > 0.30
DVARS returned by ‘fsl_motion_outliers’ is scaled to match the definition used in Power et al. (2012).
The HCP fMRI acquisitions also differ in their distribution of relative motion. In Phase I task-fMRI piloting, using a similar community sample and FSL's ‘MCFLIRT’ for estimating relative motion, we found a very similar value for the mean of relative motion across runs (0.095mm versus 0.093 mm), but with a broader distribution – i.e., a standard deviation (across runs) of the mean relative motion (within runs) of 0.11 mm versus 0.03 mm. Approximately 7.5% of runs from the Phase I task-fMRI piloting had mean relative motion values exceeding 0.20 mm, whereas only 0.5% (11 of 2207) have exceeded that level to date in the Phase II acquisitions. Notably, the Phase I piloting used a conventional acquisition (3.5 mm voxels, with a 2500 ms TR). The increased temporal resolution of the Phase II (main study) acquisitions reduces the extent of motion that can occur between consecutive time points, thus reducing relative motion estimates in motion-prone subject Whether this increased temporal resolution also confers practical benefits in terms of resistance against motion-related confounds (Power et al., 2012, Satterthwaite et al., 2012) is a topic of active investigation.
The DVARS QC metric of the FUNCTIONAL_QC pipeline is appreciably higher than has been previously reported (Power et al., 2012) because it is calculated on time series acquired at higher spatial resolution, hence, containing an increased proportion of electronic noise that have undergone only motion correction – i.e., no spatial smoothing, temporal bandpass filtering, or multiple regression of nuisance variables. Thus, no steps have been taken to reduce the amount of thermal noise in the timeseries, which leads to a mean offset of the DVARS time series from zero. This offset motivated the inclusion of the standard deviation of the DVARS time series as an additional QC measure. Despite this offset, the DVARS time series is still quite useful for visually and quantitatively identifying periods of increased motion during a run.
The nine fMRI QC measures of Table 1 are correlated to varying degrees (Table 2). In particular, tSNR, FWHM, and absolute motion all correlate with |r| > 0.68, as do mean relative motion, 95th percentile of relative motion, and percent of frames with relative motion > 0.15. A common factor analysis (using maximum likelihood factor extraction with varimax rotation) indicated a four-factor model, with the two aforementioned sets of 3 variables loading strongly on two factors (absolute value of their loadings with their respective factors > 0.68).4 These two factors explained 32% and 34% of the common factor variance (after rotation), respectively. The first factor involving tSNR, FWHM, and absolute motion is interesting. The positive relationship between absolute motion and FWHM is consistent with interpolation-related smoothing from motion correction (even though spline interpolation was used). Normally, higher smoothness would be expected to be associated with higher tSNR. However those two metrics are negatively correlated, as are tSNR and absolute motion, suggesting that absolute motion has a distinct (non-smoothness related) impact on tSNR. One possibility is that the temporal standard deviation is increased (and thus tSNR decreased) in the presence of large absolute motion due to the spatial intensity variations from the 32-channel receive coil sensitivity profile, which are not accounted for in the current FUNCTIONAL_QC pipeline. Notably, the full “fMRIVolume” pipeline (Glasser et al, this issue) does include a step to reduce the spatial inhomogeneity due to the receive coil bias field.
Table 2.
Correlations of Functional QC measures.
| tSNR | FWHM | AbsMotion | RelMotion | RelMotion 95th % | RelMotion > 0.15 | RelMotion > 0.30 | DVARS: mean | DVARS: std | |
|---|---|---|---|---|---|---|---|---|---|
| tSNR | -0.71 | -0.74 | -0.28 | -0.32 | -0.25 | -0.53 | -0.41 | -0.62 | |
| FWHM | -0.74 | 0.85 | 0.15 | 0.21 | 0.12 | 0.50 | -0.01 | 0.67 | |
| AbsMotion | -0.69 | 0.84 | 0.10 | 0.13 | 0.04 | 0.47 | -0.09 | 0.62 | |
| RelMotion | -0.37 | 0.23 | 0.16 | 0.93 | 0.95 | 0.45 | 0.40 | 0.26 | |
| RelMotion 95th % | -0.43 | 0.37 | 0.29 | 0.86 | 0.96 | 0.54 | 0.42 | 0.34 | |
| RelMotion > 0.15 | -0.29 | 0.13 | 0.05 | 0.93 | 0.72 | 0.42 | 0.41 | 0.23 | |
| RelMotion > 0.30 | -0.45 | 0.39 | 0.33 | 0.66 | 0.76 | 0.54 | 0.27 | 0.74 | |
| DVARS:mean | -0.47 | 0.03 | -0.08 | 0.53 | 0.49 | 0.50 | 0.45 | 0.18 | |
| DVARS:std | -0.54 | 0.57 | 0.52 | 0.38 | 0.63 | 0.18 | 0.59 | 0.28 |
Pearson correlations are shown below the diagonal. Spearman correlations above the diagonal. Two sets of Pearson correlations are shaded to indicate measures that loaded together strongly in a common factor analysis. N = 2207 task and resting-state fMRI runs (126 subject).
We investigated the relationship of these rapid fMRI QC metrics to task-fMRI activation in the subset of subject comprising the 20 unrelated subject of the Q1 HCP data release (Barch et al., this issue) and processed using the full “fMRIVolume” pipeline (Glasser et al., this issue). Specifically, we used the group level activation map of the 2-back versus fixation contrast of the working memory task, and the faces minus shapes contrast of the emotion task (thresholded at Z>3.29, cluster corrected) to define ROIs for computing the average Z-statistic from the first-level (i.e., run-level) activation maps for each subject and run (yielding 80 total first-level z-statistics: 20 subjects × 2 contrasts × 2 runs per contrast). There was only minimal correlation of the first-level Z-statistics with either the estimated factor scores (from the factor analysis) or the individual QC metrics (|r's| < 0.26). This demonstrates that, at least in this group of subject, the Z-statistics of individual subject activation are not strongly impacted by typical variations in subject movement. However, these 20 subjects have provided notably high quality data, and thus may not reflect the effects of more extreme levels of motion or other poor quality metrics in the first-level Z-statistics. As additional subjects are analyzed using this approach, we will be able to assess the degree to which task and resting-state output measures are related to the QC metrics in cases of more extreme subject motion. Importantly, even if the primary utility of these rapid fMRI QC metrics turns out to be flagging a small number of truly extreme outliers, the availability of these QC metrics gives us confidence that the HCP acquisitions are not being adversely affected by inconsistencies in scanner performance.
QC of Minimally Preprocessed Pipelines
The outputs of the HCP structural pipelines are examined for surface reconstruction quality in the native volume space with the native mesh using Connectome Workbench visualization software (herein referred to as ‘Workbench’; see below description of Workbench's capabilities). The efficiency of this process is increased by having a set of preconfigured Workbench ‘scenes’ that provide standardized views of the surfaces and volumes of interest. The requisite scene files are adapted to each individual subject by applying a script to a standard template scene file. An initial step is to inspect the midthickness and inflated surfaces to note for any fused sulci or missing gyri (i.e., major surface reconstruction errors). To date, these have not been encountered, suggesting that the minimal preprocessing pipelines are performing robustly. Next, the white and pial surface contours are displayed on a volume montage of T1w images to look for smaller surface segmentation or placement errors. The volume (FNIRT) and surface (FreeSurfer) registrations are checked for major errors or deformations. For the volume registration, this involves comparing the registered individual volume with the template. For the surface it involves comparing the registered “sulc” map of the individual with the atlas “sulc” map. Additionally, thresholded views of the Jacobian volume and areal distortion surface maps are viewed to check for regions of distortion that are unusually large in spatial extent. Finally, the individual myelin maps are compared with the atlas to look for any irregularities that might suggest errors in segmentation or registration (e.g., misalignment of the central sulcus to the post-central sulcus). No major errors or irregularities in the output of the structural pipeline have been identified in the subjects analyzed to date.
The outputs of the functional pipelines have been examined to ensure that single-band reference images of both phase encoding directions in MNI space are aligned with the surfaces and that the pattern of intensities on the surface of the surface-projected timeseries are reasonable (e.g., no gyral or sulcal pattern suggesting a misalignment). Because of the large number of functional runs, this QC has only been done for a subset of scans. In an initial application of these QC procedures several bugs and aspects of non-robust performance were identified and corrected in the development and early deployment of the HCP Pipelines.
QC using FIX-denoised outputs of resting state data
Resting state fMRI uses BOLD signal correlations across the brain as a measure of functional connectivity. Thus, the signal of interest in a resting state timeseries is correlated spontaneous (intrinsic) gray matter derived fluctuations. These correlations may be evaluated using seed-based correlation mapping (Biswal et al., 1995) or Spatial Independent Component Analysis (sICA, Beckmann et al., 2005). sICA separates spatio-temporal datasets into spatially independent components, each component associated with a particular timeseries. The high spatial and temporal resolution of HCP resting state data and the length of the runs (2mm isotropic, 0.72s TR, 1200 frames: 14.4 minutes) support very high dimensionality individual subject ICA of 200 components or more (Smith et al., this issue). Recently, the HCP has made use of an automated classifier of ICA components into brain-derived BOLD signal components and structured noise components. This classifier is called FMRIB's ICA-based X-noisifier (FIX). FIX has 99% sensitivity and 99% specificity in HCP resting state data when compared to human classification by experienced raters (Smith et al., this issue). Because each ICA component accounts for some portion of the timeseries variance, we can use the classifier output to apportion the timeseries variance into different categories, which can be compared across subjects or across resting state runs. In the analyses discussed below, we use the “minimally preprocessed” (including motion realignment) timeseries in grayordinate space (Glasser et al., this issue) of the publicly released 20 unrelated subjects after the addition of gentle highpass filtering (FWHM=2355s), which effectively performs a linear detrend on the data.
The simplest variance measure, TotalVariance, is the mean of the temporal variance across all grayordinates, which is then apportioned into categories according to the following model:
TotalVariance = MRVariance + MotionVariance, where MRVariance = UnstructuredNoiseVariance + StructuredNoiseVariance + BOLDSignalVariance.
The FIX cleanup routine regresses out 24 motion parameters from both the timeseries and the ICA component timeseries (Smith et al., this issue), and the standard deviation after this operation yields the overall MRVariance. The difference between TotalVariance and MRVariance is the estimated portion of the timeseries variance attributable to motion-related artifact captured by the realignment parameters, MotionVariance. Before one can determine the portion of the variance accounted for by structured noise or brain-derived BOLD, one needs to estimate the UnstructuredNoiseVariance, by regressing out all of the ICA components (both structured noise and BOLD signal) from the motion-parameter cleaned timeseries. An estimate of this variance is available because a Principal Components Analysis (PCA) dimensionality reduction, which excludes the unstructured noise, is done prior to sICA. Note that the variance categories described here are not “pure,” as ICA does not perfectly separate structured noise from Gaussian (thermal) noise (Smith et al., this issue); however, the category labels do reflect their majority constituents. Once an estimate of UnstructuredNoiseVariance is available, one can regress from the motion regressed timeseries the unique variance of the structured noise components, measure the variance, and subtract the UnstructuredNoiseVariance, producing an estimate of the BOLDSignalVariance. Similarly, StructuredNoiseVariance can be estimated by regressing the BOLD signal components out of the motion regressed timeseries, measure the variance, and subtract the UnstructuredNoiseVariance. The sum of MotionVariance + UnstructuredNoiseVariance + BOLDSignalVariance + StructuredNoiseVariance averaged about 1.26%+/-0.28% higher than the TotalVariance, suggesting that these partitioned variances are close to, but not completely independent. UnstructuredNoiseVariance is the largest component of TotalVariance, constituting 67%+/-4%, with StructuredNoiseVariance next at 16%+/-2%, MotionVariance at 14%+/-3%, and BOLDSignalVariance at 4%+/-1%. Additional useful metrics include the number of BOLD signal ICA components identified (a larger number suggests more useful information in the timeseries), the number of structured noise ICA components identified, and the total number of ICA components identified (capped at 250), the brain size (the final brain mask in native volume space as generated in Glasser et al., this issue), and the head size. An estimate of head size is generated by multiplying the T1w and T2w images in AC/PC aligned native volume space, normalizing the intensity inside the brain mask to a standard value and thresholding the image at a standard value. This produces a mask of the head that is robust across subjects.5
One notable feature of the ICA+FIX based QC metrics is their reproducibility across runs in a given subject. For each of the above metrics (plus relative motion), we performed a variance components analysis (PROC NESTED, SAS 9.2) to estimate the variability related to subject, session (nested in subject), and error (i.e., run within session) effects (Table 3). For each measure, the percent of that measure's variability attributable to subject variance exceeded 50%. UnstructuredNoiseVariance is particularly reproducible across runs, with a subject variance component of 87.9% of the total variability in this measure. The high cross-run reproducibility of the UnstructuredNoiseVariance suggests a systematic, subject-specific cause. Notably, the correlation between UnstructuredNoiseVariance and head size was r=0.88 (vs. r=0.64 with brain size). The strong correlation between UnstructuredNoiseVariance and head size is consistent with a decline in receive coil sensitivity due to increased coil loading by larger heads and likely more total ionic content in larger heads leading to increased thermal noise. In addition, UnstructuredNoiseVariance has a clear negative impact on the sensitivity for detecting resting state networks in FIX, as the correlation between number of BOLD signal components and UnstructuredNoiseVariance is r=-0.68. Both MotionVariance and StructuredNoiseVariance also have negative impacts on sensitivity for resting state networks, as the correlation between the number of BOLD signal components and these two measures is r=-0.61 and r=-0.49, respectively. Not surprisingly, MotionVariance and StructuredNoiseVariance are correlated (r=0.66), because greater subject motion leads to additional structured noise that cannot be completely regressed out with the 24 motion parameters. In the end, there is significant variability in the average number of BOLD signal components detected by ICA+FIX across subjects (mean=23.3+/-6.6 with a range of 13.75 – 38). Subjects on the low end tend to have larger head sizes and more motion variance, whereas subjects on the high end tend to have smaller heads and less motion variance.
Table 3.
Variance Components Analysis of FIX-ICA resting-state fMRI metrics.
| MeasureMeasure | Subject (%) | Session (%) | Error (%) |
|---|---|---|---|
| TotalVariance | 73.4 | 6.2 | 20.4 |
| MotionVariance | 56.1 | 7.1 | 36.9 |
| UnstructuredNoiseVariance | 87.9 | 9.1 | 3.0 |
| StructuredNoiseVariance | 69.7 | 6.0 | 24.2 |
| BOLDSignalVariance | 52.3 | 5.1 | 42.5 |
| # Total ICA Components | 84.6 | 7.1 | 8.4 |
| # Structured Noise Components | 85.0 | 6.9 | 8.1 |
| # BOLD Signal Components | 76.5 | 5.0 | 18.5 |
| RelMotion | 70.4 | 9.3 | 20.4 |
N = 80 resting-state fMRI runs (20 subjects × 2 sessions on different days × 2 runs per session). The Session effect (df=20) was nested in the Subject effect (df=19). The Error effect (df=40) represents the variability related to repeated runs within a session. For each measure, the Subject effect was significant (p<0.0004). The Session effect was significant for UnstructuredNoiseVariance (p<0.0001), # of Total ICA Components (p=0.004), # of Structured Noise Components (p=0.004), and RelMotion (p=0.04).
We also compared these ICA+FIX QC metrics to the FUNCTIONAL_QC measures discussed earlier for these same 20 subjects (80 resting-state fMRI runs). tSNR was strongly correlated with the number of BOLD signal components (r=0.73) and TotalVariance (r=-0.88), and thereby correlated with MotionVariance (r=-0.75), UnstructuredNoiseVariance (r=-0.74) and StructuredNoiseVariance (r=-0.55) (but not BOLDSignalVariance, r=0.13). Relative motion was correlated to varying degrees with those same variables (# BOLD signal components: -0.51; TotalVariance: 0.71; MotionVariance: 0.41; UnstructuredNoiseVariance: 0.64; StructuredNoiseVariance: 0.65; BOLDSignalVar: -0.06).6 Absolute motion was most strongly related to MotionVariance (r=0.78). As we extend the ICA+FIX approach to more subjects, it will become possible to analyze the factor structure of a combination of the two sets of QC metrics. Future work will seek to relate these sets of measures to each other in a larger (more robust) number of scans and to relate both sets of measures to relevant outcome-based measures of resting-state analyses, such as the sensitivity for detecting various resting-state networks. We will also explore the application of the ICA+FIX approach to HCP task-fMRI data.
QC of dMRI
QC measures for diffusion imaging (dMRI) have not yet been implemented. This delay is, in part, attributable to the complexity of the HCP dMRI processing stream, which accounts for imaging gradient non-linearities both in diffusion sensitization and spatial encoding, as well as eddy current effects (Sotiropoulos et al., this issue; Ugurbil et al., this issue). In fact, GPU-assisted computation is required to achieve practical throughput, and this implementation is currently nearing completion. We anticipate that QC measures for diffusion imaging (dMRI) will be available within the next year.
QC of non-imaging data
The HCP protocol includes several non-imaging data types, including two extensive behavioral batteries, intake questionnaires, and a 7-day retrospective survey of alcohol and tobacco use that is taken at the end of the participant's visit. The NIH Toolbox and Penn computerized battery used for behavioral testing include built-in quality control checks to ensure that consistent data are captured as the batteries are administered. The data generated by these systems are automatically imported into IntraDB to minimize labor and data entry errors. The various intake and exit data are entered directly into IntraDB, which includes field-level value checks that require explicit user overrides to allow out of range entry. The subject visit details are reviewed daily by the study coordinator to ensure completion of data entry, and import and consistency of subject identifiers across study domains. The full non-imaging data is spot checked on a monthly basis to ensure overall completeness and accuracy.
ConnectomeDB
Organization of the database
Several organizational aspects of ConnectomeDB are worth noting. First, the data are grouped into XNAT projects that match the HCP quarterly data release schedule. Throughout each quarter as data are collected, once they have passed QC the data are pushed from IntraDB to a “private” project that is accessible only to internal HCP staff. Maintaining these projects in “private” mode allows processing pipelines and other preparatory tasks to be executed and reviewed prior to open access public release. When the release date arrives, the project is switched to “public” mode, which instantaneously makes the data accessible to all users who have agreed to the HCP open access data use terms. In addition to facilitating the data preparation process, maintaining separate projects for each quarterly release provides a user friendly mechanism for highlighting recent data sets and enables users to download and explore just the new data.
Second, the imaging data, which have been acquired over four or five individual imaging sessions for each participant, are merged during the transfer from IntraDB to ConnectomeDB into a single “mega” session, which simplifies the data structure for end users. The merge process involves an extensive set of rules for determining which scans to include, their sequential order, and labeling. Structural scans marked as “fair” or “poor” by QC reviewers, for example, are excluded, as are fMRI scans that lack 25% or more of the expected number of frames. The interdependencies between scans by shim value and associated bias and fieldmap scans are identified and explicitly tagged in the database. These tags are subsequently used by the processing pipelines and to organize files for distribution. In addition, a document is generated and written to the structural data packages, both unprocessed and preprocessed (see below) to assist users in independently processing the data.
Third, much effort has gone into ensuring that all instrument-generated testing dates have been redacted from the data, consistent with HIPAA privacy protections. In addition, the voxels around faces and ears in high resolution structural scans have been blurred (“defacing”), and subject ages have been binned in 4-5 year bands to reduce their precision. A number of fields representing particularly sensitive data (e.g., history of drug use) or data that in combination might be identifying, such as race, body weight, age by year, and/or family status (e.g., identical twins) are excluded from the open access release. These data are available only to qualified users who agree to the HCP's Restricted Access Data Use Terms, which involve important constraints on how this information can be published (Van Essen et al., this issue). This dual open/restricted access approach has been vetted by the Washington University Human Research Protection Office and HIPAA Privacy Offices. An independent consultant with extensive experience in protecting the privacy of family study participants has also determined that HCP's approach is appropriate to optimizing scientific utility while ensuring subject privacy.
Automated image processing
All of the image processing routines in use by the HCP are being implemented as automated pipelines within the HCP infrastructure using the XNAT Pipeline Service (Marcus et al., 2007) (Table 4). To date, the automated QC for fMRI and “version 1.0” of the minimal preprocessing procedures for all modalities (Glasser et al., this issue) have been completed and run on each subject. Pipelines are launched either in batches at the command line or by operations staff from the database web interface, depending on the particular processing context. Pipelines execute on one of two computing systems accessible to the HCP. A cluster with 64 dedicated cores and 140 shared cores is used to execute pipelines that have modest computing requirements but need to be completed with short latency on demand, including defacing, protocol validation, and quality control. A high performance computing system managed by the Washington University Center for High Performance Computing is used to execute pipelines that are computationally demanding but have flexible scheduling demands, including all of the preprocessing pipelines. All files generated during pipeline execution are posted to ConnectomeDB. Subsets of these files are then packaged using the Packaging Service described below in preparation for sharing. The outputs of the structural pipelines are mandatory for other modality pipelines that register outputs to MNI and native structural space (see Glasser et al., this issue).
Table 4.
Automated image processing pipelines.
| Name | Description | Products | Core Methods | Run Time |
|---|---|---|---|---|
| Protocol validation | Inspects DICOM headers to verify image acquisition | Validation Report | XNAT | 1m |
| Defacing | Blurs areas of structural images containing facial features. | DICOM w/ modified voxels | Milchenko & Marcus | 1m/series |
| NIFTI conversion | Generates NIfTI-formatted versions of acquired images | NIFTI | dcmtonii | 5m/series |
| Phantom QC | Calculates RMS stability, drift, mean value, and SNR | QC Report | BIRN | 15m |
| fMRI QC | Calculates mean, variance, skewnewss and kurtosis of DVAR | QC Report | FSL | 15m |
| Structural MR QC | Detects blurring, edge coherence, and SNR | QC Report | python | 5m |
| Pre-surface generation | Removes spatial artifacts and distortions from structural scans, co-registers them to common atlas space. | Undistorted native and MNI volumes | FSL/python | 8h |
| Surface generation | Generates cortical surfaces and segmentations. | Native Surfaces, Segmentations | Freesurfer, FSL | 24h |
| Post-surface generation | Generates Workbench-ready data files and myelin maps. | Workbench spec files | Workbench,FSL | 4h |
| fMRI volumetric processing | Removes spatial distortion, motion correction, and co-registers structural scans to common atlas space. | MNI registered fMRI volumes | Freesurfer, FSL, python | 4h |
| fMRI surface processing | Maps fMRI volume timeseries to surfaces and creates standard grayordinates space. | CIFTI dense timeseries in grayordinates | Workbench, FSL | 4h |
| Diffusion* | Intensity normalization, EPI distortion removal, eddy-current distortion removal, motion/gradient-nonlinearity correction, structural registration | Diffusion Data in Structural Space | FSL, Freesurfer, python | 36h |
| Fiber Orientation Modeling* | Bayesian estimation of diffusion parameters obtained using sampling techniques | Probabilistic diffusion orientations | FSL | TBD |
| MEG/EEG* | TBD | TBD | Fieldtrip | TBD |
| Connectivity* | TBD | TBD | Workbench/FSL | TBD |
| FIX* | Run ICA and classify ICA components with removal of noise-based components | rfMRI Denoising | FSL/matlab/R | 14h |
| Task fMRI Subject Analysis* | Single subject task fMRI level 1 and 2 GLM analysis | Workbench/FSL | 20m |
The HCP's pipelines are implemented using XNAT's Pipeline Service. Run time is shown in minutes based on execution on a XXX system. Pipelines denoted with an asterisk are currently in development and run times are estimated
Data versioning strategies
As advances are made to the HCP's processing and analysis methods, the HCP's pipelines will be updated. Modified versions of files and derived data generated by the pipelines will be uploaded to ConnectomeDB. In an ideal world, all previous versions of the files would continue to be made accessible as in a typical software version control system. However, due to the scale of the connectome data, we cannot guarantee that historical versions of the data will be preserved indefinitely. Several steps are being taken to mitigate the impact of this process. First, the HCP recommends that investigators preserve on their own any data they use in analyses they conduct locally. Second, while the data produced by pipelines is not under version control, the pipeline code and software upon which the pipelines depend is under full version control. Should a researcher desire to reconstruct a historical version of the data, the code could be extracted from the version control system and run against the unprocessed data to generate an effective equivalent of the historical data (though due to minor differences in compilers and computing platforms, the recreated data is unlikely to be an exact numeric copy.) Third, the data files in ConnectomeDB and packages downloaded from it are tagged with the version of the pipeline and execution run used to generate them. The execution run defines the specific parameter settings that may be varied while maintaining the same software version. Finally, all services implemented on ConnectomeDB will produce a “receipt” indicating the version of the service and the version of the data against which it was run.
Data sharing strategies
The data set generated by the HCP, including the acquired images, minimally preprocessed data, and full dense connectomes, is expected to approach 1 petabyte. Given this massive volume, it is unrealistic to expect users to simply download the data from a website. Instead, we have taken a multi-pronged approach to match a specific data sharing mechanism to the specific context. For full data sets of a small number of subjects, ConnectomeDB includes network-based services for data transfers. For data sets of a large number of subjects, the HCP will ship (at cost) hard drives (“Connectome in a Box”) pre-populated with data organized by the HCP's quarterly release schedule. The threshold for when a data request is best met by a network transfer versus a hard drive shipment depends on the bandwidth capacity of the user and the current demand on ConnectomeDB. This threshold may also change over time as overall Internet bandwidth expands (see Downloading Data below). Presently, the HCP recommends that users request no more than 20 full subject data sets at a time. The details of the ConnectomeDB and Connectome in a Box systems are described below.
In addition, the HCP is implementing computational services within ConnectomeDB so that analyses can be conducted without the need for transferring the data. Indeed, bringing algorithms to the data rather than data to the algorithms is the most efficient option for connectome-scale data. An initial proof of concept service has been implemented that calculates average connectivity across an arbitrarily chosen group of subject for an individual grayordinate. For a connectome of approximately 90,000 ‘grayordinates’ (vertices and voxels, see below) and a group size of 20 subjects, this calculation executes in less than 1 sec on the ConnectomeDB infrastructure. This proof of concept implementation illustrates that rich data exploration services can be performed in real time as users navigate the data. Programmable interfaces to these services will be made available via REST web services API, which can be accessed by any variety of client applications, including by ConnectomeDB itself, by Connectome Workbench, as described below, and by tools created by developers outside the HCP. While the formal API structure for these services if still in development, we expect each service request will include several standard parameters including one or more subject groups and one more regions of interest (ROIs) over which the requested service would operate. Currently, a preliminary list of potential services is being drafted to help scope the overall requirements and includes services to compute: an n × n correlation matrix for arbitrary group of n ROIs supplied by the user, server returns n × n correlation matrix; summary statistics on behavioral measures over a subject group, with adjustments for familial structure as required; locations of activation in t-fMRI reported by peak coordinates, radius, and size in voxels; locations of activation in t-fMRI, reported by parcellation region; correlation over a subject group between behavioral measures and t-fMRI activation.
Data Dictionary
The HCP behavioral battery and study metadata include hundreds of individual data fields. To communicate the meaning of each of these terms to users, a data dictionary service was implemented on ConnectomeDB. This service comprises three components: a document containing the actual terms, their definitions, and an assortment of helper content; an API for querying the database for dictionary entries; and user interface elements for viewing and navigating the dictionary. The dictionary document includes a hierarchical structure with separate levels for categories, assessments, and individual data fields. The dictionary service enables client tools, such as the web interface, to query ConnectomeDB for project-specific dictionary details at each level of the dictionary. The service returns the portion of the dictionary, currently formatted as JSON, that matches the query. The dictionary user interface (UI) component is primarily manifested on the Data Dashboard (see below), where it is used to generate filters for searching the data. Ongoing work on the dictionary service aims to implement links to formal ontologies such as the Cognitive Paradigm Ontology (Turner and Laird, 2012) and to expand the user interface components to enable review of the full dictionary and to cover more of the overall ConnectomeDB website.
Data Packages
The HCP preprocessing pipelines generate a large number of files, many of which are not essential for typical analyses but are retained in the database for quality control and review purposes. To streamline distribution of the high interest files, a file package generation service was developed that allows specific subsets of the files hosted in ConnectomeDB to be compiled into Zip-formatted distributable file archives. The service uses specification documents to define the contents of a particular package based on file naming patterns along with other metadata. Packages are typically defined to include contents for a single subject, but group level data can also be packaged. Currently, a script external to ConnectomeDB itself is used to pre-generate and store packages. The next generation of this service, which is currently in development, will dynamically assemble and stream the packages on demand, eliminating the inefficient storage of the pre-generated packages and allowing a larger number of package configurations, including user-defined custom packages.
Along with the data contained in the download packages, the download service creates a checksum of the generated download package. This provides validation that the download was completed successfully without errors during the transfer process. In addition, the download package itself contains a download manifest, which has information about each of the resources within the download package, including a checksum for each file. Along with dynamically generated download packages, future releases will include a transfer manager application that can be used to manage local file repositories. The download manifest in these generated packages can be imported into the transfer manager, which will allow users to update only those files that are new or have changed since a previous download.
User Interface
Basic Navigation
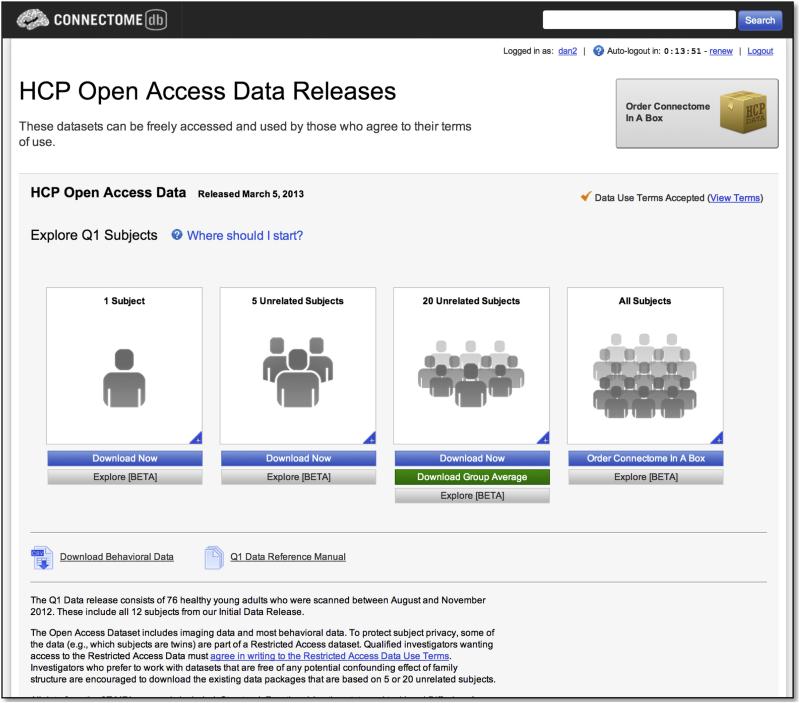
ConnectomeDB includes a modified version of the standard XNAT navigation structure. A landing page details the available data and guides the user through accepting data set-specific data use terms (Figure 5). The page is designed to highlight recent releases, such as the initial open access “Q1” HCP data release in March, 2013. Once the user agrees to the open access terms for a data set, s/he is automatically granted access to the open access components of the data. For the Q1 release, links are provided to pre-selected groups of 1, 5, and 20 unrelated subjects that are known to have full high quality data acquisitions and that have been used extensively within the HCP consortium in early analyses and thus serve as good reference data. This view also helps manage network bandwidth limitations by encouraging users to download only as much data as they need to accomplish their current goals. For each of these groups, as well as for the full data set, the user can follow links to the Data Dashboard screen, which provides a searchable view of all subject metadata, and the Package Download screen, which provides a streamlined interface for selecting and downloading pre-packaged file archives for one or more modalities. Users can also access the standard XNAT reports and navigation elements to explore individual subjects. These pages have also been customized to reflect the specific metadata and data organization of the HCP.
Figure 5. Basic navigation in ConnectomeDB.
The landing page guides users to different pre-selected data sets and to the full Q1 open access data release. The pre-selected groups were specifically identified to provide user friendly access to various subject group sizes, all containing high quality data from unrelated subjects.
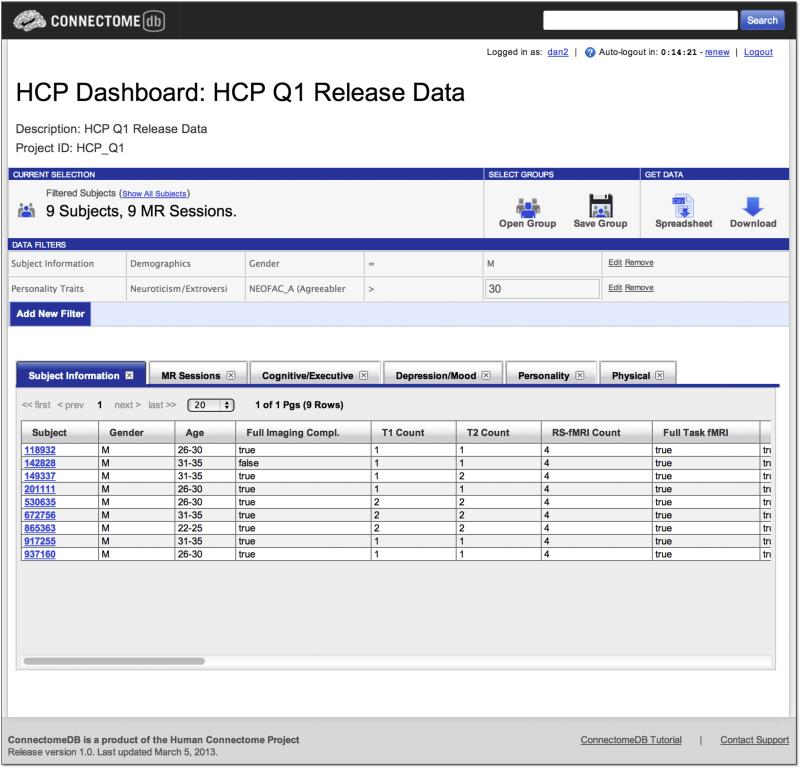
Data Dashboard
ConnectomeDB's Data Dashboard provides an interface for dynamic exploration of the HCP dataset (Figure 6). Interactions on the dashboard are built around the concept of subject groups – subsets of the HCP data set who match some specified criteria. The dashboard includes several components for interacting with subject groups: a tool ribbon provides access to various actions, including saving subject groups, opening previously saved groups, and downloading data for the current subject group; data filters for defining the subject group criteria; and a tabular display of demographics, behavioral measures, subject data and metadata for the currently selected group. The dashboard components use the data dictionary, with the filter selectors and tabular data displays mapping directly to the dictionary hierarchy elements and the data tables drawing from the dictionary categories. Future versions of the dashboard will enable execution of the various computational services described above for the selected subject group and direct visualization of the results on an interactive 3D surface model.
Figure 6. Data Dashboard.
The Data Dashboard allows users to filter the data set by any of the metadata and data fields in the database. Here, the user has selected all male subjects with an agreeableness score on the Neuroticism-Extraversion-Openness Inventory greater than 30. The results set of 9 subjects is displayed in the data tables, with tabs for the each of the data categories. The user can then choose an action (download a spreadsheet, download images) for the subject group they have generated.
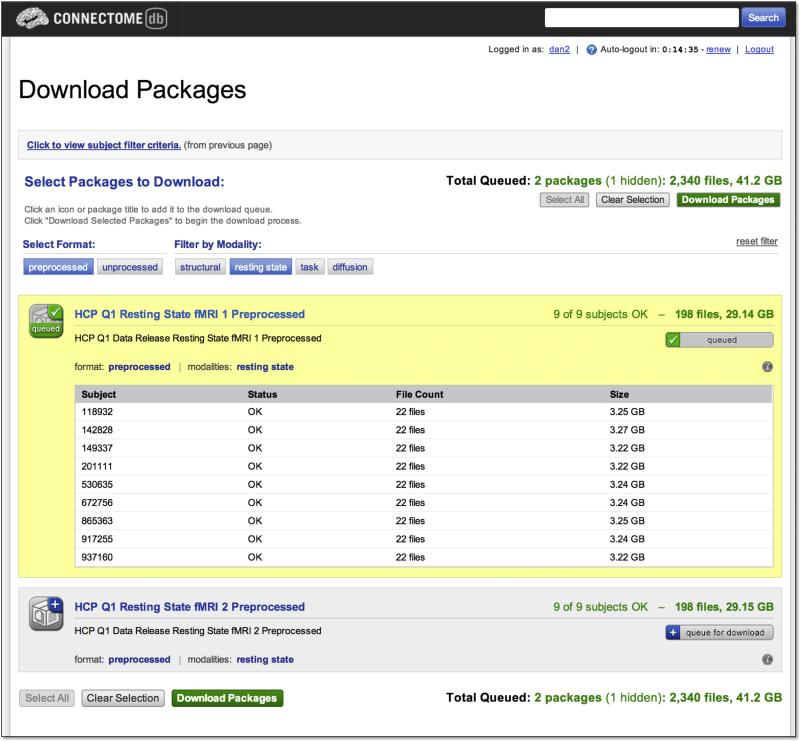
Downloading data
ConnectomeDB's Download Packages interface utilizes the Data Package service described above to enable users to download individual subject or subject group data selected generated through the Data Dashboard (Figure 7). The interface presents users with a filterable list of packages for unprocessed and preprocessed version of each of the modalities included in the HCP protocol, including the structural, diffusion, fMRI resting state, and each fMRI task. The unprocessed data packages include NIfTI-formatted files both the left-to-right and right-to-left phase encoded acquisitions for the selected modality and repeat acquisitions when relevant. For each package type, details of the package contents, including number of files and total size, are displayed along with information about the availability of the files for each subject in the selected subject group.
Figure 7. Download Packages.
The Download Packages interface allows users to filter the set of packages to download by format and modality and to review the data that will be downloaded. The interface automatically uses the subject group previously selected by the user, in this case the 9 subjects generated in the search illustrated in Figure 6. After queuing the packages of interest, clicking the “Download Packages” launches the Aspera Connect client to execute the actual download.
Once the set of packages has been selected, the user is directed to a download interface that uses the commercial Aspera fasp[TM] high speed data transfer technology(Aspera, 2013). The connection-based Internet standard protocol TCP is notoriously inefficient over high-bandwidth, high-latency routes (“long fat networks” or LFNs; Lakshman and Madhow, 1997). This inefficiency limits the usefulness of TCP-based protocols such as FTP and HTTP for moving big data across LFNs. A variety of techniques have been used to compensate for TCP's LFN problem, including extensions to the TCP protocol and multiple simultaneous TCP connections (Hacker and Athey, 2001). Aspera's fasp[TM] is a proprietary transport built on the datagram-oriented Internet standard protocol UDP and designed to maximize throughput across LFNs.
Aspera's fasp supported 100 Mbit/s transfers to multiple simultaneous users within a university network. Single download testing from remote sites yielded steady transfer rates of 70-85 Mbit/s between university networks in the US and UK. For comparison, over standard FTP, these same sites obtained transfer speeds of 4-12 Mbit/s. Based on these tests, downloading a typical 10 GB single subject unprocessed HCP data set to a typical US university should take less than 15 minutes over fasp compared to nearly 3 hours over FTP.
Connectome in a Box
Owing to the massive size of the HCP data set, downloading large portions of the data over the Internet is at the limits of current network bandwidth and latency constraints, particularly for international locales. As an alternative to network-based downloads, we have developed a physical data transport mechanism that we refer to as Connectome in a Box (CBox). CBox is based on high capacity (currently 3 TB) consumer grade hard drives on which the HCP unprocessed and preprocessed data are loaded. Copies of CBox formatted for Linux, Windows, and Mac systems can be ordered via the HCP website along with an optional external drive enclosure. The CBox copies are generated on a custom-built duplication system that is capable of generating up to 15 simultaneous copies of a master instance using the open source Clonezilla package. The initial data release, which has a capacity of 2.0 TB, requires 10 hours to duplicate on the system, with an additional 10 hours to verify replication accuracy. The drives are delivered at cost to customers via FedEx. Users can attach the drive directly to their laptop or workstation via a USB or eSATA connector and work directly from the shipped drive or transfer the data to alternative storage, such as communal network storage hosted by a department or an imaging center. Communal storage has the advantage of serving as a single access point for multiple investigators at a center; but all investigators accessing the data are expected to sign and abide by the HCPs data user terms. CBox data are organized in the same directory structure as data downloaded directly from ConnectomeDB, but is pre-extracted for the end user's convenience.
Connectome Workbench
Connectome Workbench (henceforth “Workbench”) is a surface and volume visualization platform developed by the HCP for viewing the many modalities of MRI-based data generated by the HCP minimal preprocessing and analysis pipelines. It is freely available (http://www.humanconnectome.org/connectome/connectome-workbench); the beta 0.8 version (March, 2013, release) is accompanied by a tutorial and associated dataset.
Workbench supports standard neuroimaging NIfTI and GIFTI data formats plus the recently introduced CIFTI format for ‘grayordinate’ representation of surfaces and volumes compactly in a single file (http://www.nitrc.org/projects/cifti/; see below). Workbench also includes many command-line utilities (“wb_command) that perform a wide variety of algorithmic tasks using volume, surface, and grayordinate data; these utilities are extensively used in the HCP minimal preprocessing pipelines (Glasser et al., this issue). Workbench and wb_command are freely available C++ binaries that are compiled to run on Windows 64-bit, Windows 32-bit, Mac OS X 64-bit, and Linux 64-bit operating systems. Workbench is a successor to Caret5 (Van Essen et al., 2001), with extensive revisions to increase performance, improve interface design, and enable easier and more flexible viewing of multi-modal imaging data.
Workbench enables viewing cortical surface data on individual hemispheres or in “montage” views that concurrently display left and right hemispheres with 180-degree opposing views (e.g., medial and lateral, see Fig. 8, left). Volume data can be viewed as individual slices, as a montage (or “lightbox”) of many slices (Fig. 8, right), or as three orthogonal slice places. An oblique slice viewing mode is also planned. Additionally, surface and volume data can be viewed simultaneously in a Whole Brain viewing mode that allows concurrent display of surface models, volume slices, and 3D volume voxels. In any of the volume views, cortical surfaces can be viewed on volume slices as surface contours (with or without scalar data mapped onto the contour), as shown in Fig. 8 (right). The major forms of data that can be viewed on the surface or volume are scalar maps (containing real-valued data mapped to a color palette, such as a cortical myelin map) and label maps (containing specific colors and names for integer values, such as a parcellation of cortical areas and subcortical nuclei). Multiple maps can be overlaid as layers, each with its own color palette and threshold settings. Maps can be easily turned on and off, added, removed, or reordered, or have their opacities modified to facilitate quick comparisons. Additionally, Workbench can be used to create and display borders (boundaries projected onto the cortical surface, such as the outline of an ROI) or foci (points, such as the centers of gravity of a task activation cluster).
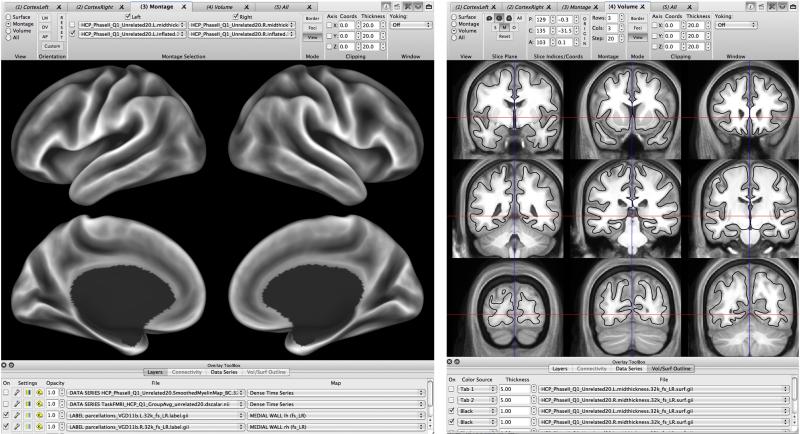
Figure 8. Example of surface and volume visualization in Connectome Workbench.
Left: A montage view of a population-average inflated surface showing the FreeSurfer-generated average ‘sulc’ map displayed on medial and lateral views of left and right hemispheres. Right. A montage volume-slice view of the population-average T1w volume. The population-average midthickness surface (black contour) does not perfectly overlay the average T1w gray-matter pattern, because there are significant differences in the inter-subject alignment achieved by nonlinear volume-based vs surface-based registration.
Workbench makes extensive use of CIFTI files (Glasser et al., this issue) to represent both cortical surfaces (vertices, with topological relationships encoded) and subcortical grey volume data (voxels) in a single data structure. Workbench determines from the CIFTI header which brain structure and location on which to display the data (e.g. left cortical surface vertex 1000 or right thalamus voxel coordinates 3.9, -1.4, 10.6). Workbench supports multiple CIFTI file formats that represent complementary data types. A dense timeseries CIFTI file (grayordinates × time) can be viewed as a series of maps displayed on cortical surfaces and subcortical volume slices or as a timecourse for a selected grayordinate, using the timecourse graph window (and as an average timecourse for all grayordinates within a selected ROI). A dense scalar file (grayordinates × scalar maps) can be viewed using the same options, but differs insofar as it contains a series of spatial maps (e.g. tfMRI contrast maps or ICA component spatial maps) that can have distinct labels for each frame. Dense label files (grayordinates × label maps) based on combined cortical and subcortical parcellations are also supported. A dense connectome file (grayordinates × grayordinates) contains a connectivity value between every grayordinate and every other grayordinate. Because dense connectivity files can be very large (~32.5 GB for HCP-generated dense connectomes), it is noteworthy that they can be accessed interactively by reading out only one row at a time or an average across a user-selected subset of rows from a file that is stored remotely (e.g., in the ConnectomeDB database, as described above). This enables the user to click on a grayordinate (or draw an ROI of grayordinates) and instantaneously view the map that corresponds to those grayordinates’ connectivity with every other grayordinate. These maps can either represent functional connectivity (based on Pearson correlation of resting-state timeseries data) or structural connectivity (based on tractography).
Dense connectomes need not be square matrices of the same dimensions. For example, a grayordinates × “whiteordinates” (white matter voxels) map can display the spatial probability distributions of tractography results associated with each grayordinate. Taking this idea a step further, a dense trajectory file stores not only the number of tractography samples in each white matter voxel, but also up to three modeled fiber orientations. This enables visualization of fiber trajectories similar to deterministic streamlines, while preserving the probabilistic nature of the tractography data by weighting the opacity of the fiber orientations according to the probability that they are associated with a selected seed location (Sotiropoulos et al., this issue). These trajectories can be visualized in Workbench either in volume slices or in the whole brain view, together with cortical surfaces that can be opaque or translucent.
Workbench also supports visualization of parcellated connectomes. A parcellated Connectome contains connectivity relationships between each cortical or subcortical parcel and every other parcel. This large dimensionality reduction greatly reduces file sizes. Parcellated connectomes can also contain a dense dimension (e.g. parcels × dense or dense × parcels), which may be useful for certain applications. In the future, parcellated timeseries may be useful for ROI-based tfMRI, rfMRI, or EEG/MEG analyses (see Larson-Prior et al., this issue). In general, the utility of parcellated connectome datasets depends on the quality of the parcellations, which is a domain of active investigation both within and outside the HCP consortium.
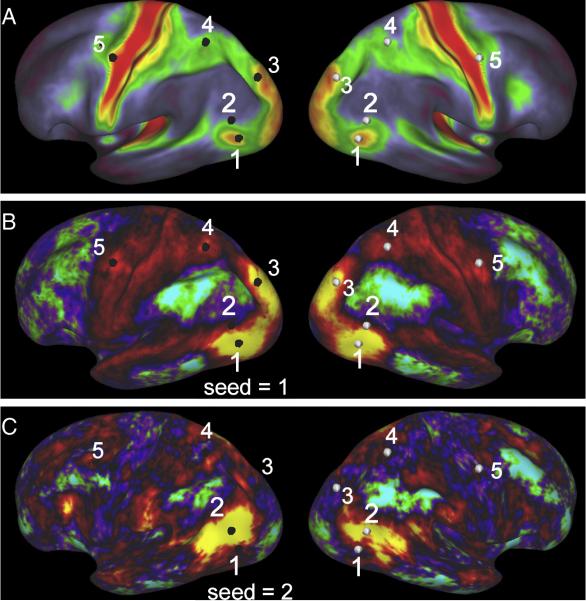
The following example illustrates key aspects of the wide range of visualization options available in Workbench, with its multiple viewing windows and multiple browser-like tabs within each window. Suppose a user is interested in the relationship of data pertaining to major HCP modalities in and around the heavily myelinated visual area complex known as MT+. One window can be used to display myelin maps (Fig. 9A), and another window (or tab) to display resting-state functional connectivity maps whose surfaces are yoked to present identical views (Fig. 9B, C). Clicking in the center of MT+ based on heavy myelin content (vertex 1 in Fig. 9A) reveals functional connectivity for that vertex in the both left and right hemispheres (Fig. 9B). The additional highlighted vertices facilitate comparisons of correspondences between hotspots of functional connectivity and heavy myelination in both the left (black vertices) and right (white vertices) hemispheres. Clicking on a vertex just dorsal to MT+ (vertex 2 in Fig. 9A) reveals a very different map of functional connectivity (Fig. 9C) whose hotspots lie mainly in regions of low or moderate myelination. Additional windows or tabs could be used to display task-fMRI (tMRI) contrasts, tractography-based connectivity maps, and the differing 3D trajectories taken by probabilistic tractography streamlines emanating from inside and outside MT+. Finally, the user might in each window include layers representing multiple individuals for each modality as well as a group average, taking advantage of the one to one correspondence established across subjects using the standard grayordinates space (Glasser et al., this issue).
Figure 9. Multi-modal comparisons in Connectome Workbench.
A. Group-average myelin map, with 5 surface vertices highlighted on the left hemisphere (black) and geographically corresponding vertices displayed on the right hemisphere (white). B. Functional connectivity map for vertex 1, centered on MT+ in the left hemisphere. Many hotspots of functional connectivity are centered on regions of heavy myelination. C. Functional connectivity map for vertex 2, situated just dorsal to MT+. The pattern of functional connectivity is very different and is largely centered in regions of low or moderate myelination. To reduce complexity, the medial views of each hemisphere visible in Workbench montage view are not illustrated here.
Complicated sets of windows and layers of the type illustrated in Fig. 9 generally take considerable time to set up. However, this need only be done once, because Workbench supports saving of “scenes” that store all the information needed to regenerate complex scenes in their entirety, even when such scenes access data that are a mixture of files stored locally and in ConnectomeDB. Not only does this greatly reduce setup time when returning to an analysis later, but it also can facilitate collaborations among investigators by transmitting appropriate scene files along with the associated datasets.
Workbench remains under active development by the HCP, and many additional capabilities are anticipated in the next several years. One such feature will be web-enabled specification (‘spec’) files. A spec file is a standard set of data files that the user wants to open together. It is similar to a scene file, except it does not specify how the loaded data are to be displayed. If a user accessed a spec file located in ConnectomeDB (via Workbench using the appropriate URL), Workbench could then download all of the data and display them locally for the user. A similar process will also be implemented for scene files, so that the database can generate a spec or scene file based on a user query. The file could then be downloaded and explored by the user in Connectome Workbench. Thus scene files and spec files will serve as an integral link between ConnectomeDB and Connectome Workbench for data visualization and data mining.
Discussion
The primary data acquisition and first order quality control procedures for the HCP are stable. However, the many second order quality control procedures, informatics tools, and data mining capabilities described here reflect a project still in very active development and evolution. For example, as more subjects are studied, we expect to develop a better understanding of the various quality metrics and to potentially establish cutoff points for acceptance in analyses. Much of the current focus pairs with ongoing development of image processing and analysis methods by other components of the HCP. As these reach a reach a level of readiness, they will be incorporated into ConnectomeDB's automated pipelines and computational services infrastructure. Specific pipelines likely to be impacted soon include diffusion quality control, MEG preprocessing, and dense connectome generation.
IntraDB and ConnectomeDB include a variety of customizations on the XNAT system, many of which are general and reusable in nature. These include project-specific data user agreements, the dictionary service, the data dashboard, the package service, the download interface and API, the IntraDB to ConnectomeDB transfer service, the integration of Aspera fasp, the various QC and preprocessing pipelines, and the data models for the various behavioral assessments used by the HCP. These custom features are being extracted as pluggable modules that can be downloaded from XNAT Marketplace and deployed on independent XNAT systems. Several additional informatics components are not reusable per se but provide novel approaches to data sharing, including the Connectome in a Box distribution mechanism and data organization strategies.
In addition to these specific modules, the overall code for the HCP databases is open source and available on the Bitbucket version control system (https://bitbucket.org/hcp). The software may be used by projects conducting similar research as the HCP, though significant modifications to the code would likely be necessary to meet each project's specific requirements.
The HCP has promoted the co-evolution of a database (ConnectomeDB) and a visualization platform (Workbench) that each has extensive free-standing capabilities. Currently, they are linked by entry-level capabilities that involve Workbench's ability to remotely access dense connectome datasets in ConnectomeDB. We consider this a valuable toehold into a much more intricately intertwined set of data mining capabilities envisioned for the future. This will enable interactive exploration and visualization of a wide variety of complex relationships between brain circuits, brain function, behavior, heritability, and their genetic underpinnings, based on the rich and systematically acquired and analyzed datasets made available through the HCP.
Highlights.
Quality control procedures of the HCP have enabled high throughout data acquisition.
IntraDB and ConnectomeDB database services enable operations and open access data sharing.
Connectome Workbench enables cross-modal data visualization and exploration.
Acknowledgements
The authors would like to acknowledge the contributions of the many investigators and staff on the WU-Minn Human Connectome Project and the participants who have made it possible. This work was supported in part by the NIH through the following grants: NIH 1U54MH091657, funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University; NIH: U54 MH091657, U24 GM104203, and R01 EB009352.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
In some of the early Phase II structural sessions, the shim currents in effect during the gradient echo fieldmap acquisition differed slightly from the main structural (T1w and T2w) scans because of differing FOVs between those scans and the fact that automated copying of the “Adjust Volume” to the fieldmap scan had not yet been set in the protocol.
Shorter TR reduces static image SNR due to reduced recovery of longitudinal magnetization.
The third factor included strong loadings (> 0.5) from 95th percentile of relative motion, percent of frames with relative motion > 0.30, and standard deviation of the DVARS time series. The fourth factor was primarily loaded (0.93) by just the mean of the DVARS time series.
Because the T1w and T2w images have undergone defacing, the head size estimate may be different from the non-defaced images.
tSNR and relative motion were themselves correlated (r=-0.58) in this subset of 80 scans (vs. r=-0.37 in the larger set of scans used for Table 2).
References
- Aspera [March 11, 2013];Aspera fasp High Speed Transport. 2013 at http://asperasoft.com/fileadmin/media/Asperasoft.com/Resources/White_Papers/fasp_Critical_Technology_Comparison_AsperaWP.pdf.
- Barch DM, Burgess G, Harms MP, Petersen SE, Schlaggar BL, Corbetta M, Glasser MF, Curtiss SW, Dixit S, Feldt C, Nolan D, Bryant E, Hartley T, Footer O, Bjork JM, Poldrack R, Smith S, Snyder AZ, Van Essen DC. Function in the human connectome: Task-fMRI and individual differences in behavior. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.05.033. (Special issue on Mapping the Connectome) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philosophical Transactions of the Royal Society B: Biological Sciences. 2005;360(1457):1001. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Bodurka J, Ye F, Petridou N, Murphy K, Bandettini PA. Mapping the MRI voxel volume in which thermal noise matches physiological noise--implications for fMRI. Neuroimage. 2007;34:542–549. doi: 10.1016/j.neuroimage.2006.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Glover GH. Report on a multicenter fMRI quality assurance protocol. J Magn Reson Imaging. 2006;23:827–839. doi: 10.1002/jmri.20583. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson T, Fischl B, Andersson J, Xu J, Jbabdi S, Webster M, Polimeni J, Van Essen DC, Jenkinson M. The minimal preprocessing pipielines for the Human Connectome Projects. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.04.127. (Special issue on Mapping the Connectome) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover GH, Mueller BA, Turner JA, Van Erp TGM, Liu TT, Greve DN, Voyvodic JT, Rasmussen J, Brown GG, Keator DB, Calhoun VD, Lee HJ, Ford JM, Mathalon DH, Diaz M, O'Leary DS, Gadde S, Preda A, Lim KO, Wible CG, Stern HS, Belger A, McCarthy G, Ozyurt B, Potkin SG. Function biomedical informatics research network recommendations for prospective multicenter functional MRI studies. Journal of Magnetic Resonance Imaging. 2012;36:39–54. doi: 10.1002/jmri.23572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve DN, Mueller BA, Liu T, Turner JA, Voyvodic J, Yetter E, Diaz M, McCarthy G, Wallace S, Roach BJ, Ford JM, Mathalon DH, Calhoun VD, Wible CG, Brown GG, Potkin SG, Glover G. A novel method for quantifying scanner instability in fMRI. Magn Reson Med. 2011;65:1053–1061. doi: 10.1002/mrm.22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur RC, Ragland JD, Moberg PJ, Turner TH, Bilker WB, Kohler C, Siegel SJ, Gur RE. Computerized neurocognitive scanning: I. Methodology and validation in healthy people. Neuropsychopharmacology. 2001;25:766–776. doi: 10.1016/S0893-133X(01)00278-0. [DOI] [PubMed] [Google Scholar]
- Gur RC, Richard J, Hughett P, Calkins ME, Macy L, Bilker WB, Brensinger C, Gur RE. A cognitive neuroscience-based computerized battery for efficient measurement of individual differences: standardization and initial construct validation. J Neurosci Methods. 2010;187:254–262. doi: 10.1016/j.jneumeth.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker TJ, Athey BD. The End-to-End Performance Effects of Parallel TCP Sockets on a Lossy Wide-Area Network.. 16th IEEE-CS/ACM International Parallel and Distributed Processing Symposium; Ft. Lauderdale, FL IEEE-CS/ACM. Apr. 2002.2002. [Google Scholar]
- Helmer KG, Ambite JL, Ames J, Ananthakrishnan R, Burns G, Chervenak AL, Foster I, Liming L, Keator D, Macciardi F, Madduri R, Navarro J-P, Potkin S, Rosen B, Ruffins S, Schuler R, Turner JA, Toga A, Williams C, Kesselman C. Enabling collaborative research using the Biomedical Informatics Research Network (BIRN). J Am Med Inform Assoc. 2011;18:416–422. doi: 10.1136/amiajnl-2010-000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton C, Josephs O, Stadler J, Featherstone E, Reid A, Speck O, Bernarding J, Weiskopf N. The impact of physiological noise correction on fMRI at 7 T. Neuroimage. 2011;57:101–112. doi: 10.1016/j.neuroimage.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, L Whitwell J, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DLG, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshman TV, Madhow U. The performance of TCP/IP for networks with high bandwidth-delay products and random loss. IEEE/ACM Trans. on Networking. 1997;5(3):336–350. [Google Scholar]
- Larson-Prior LJ, Oostenveld R, Della Penna S, Michalareas G, Prior F, Babajani-Feremi A, Marzetti L, de Pasquale F, Di Pompeo F, Stout J, Woolrich M, Luo Q, Bucholz R, Fries P, Pizzella V, Romani GL, Corbetta M, Snyder AZ. Adding dynamics to the Human Connectome Project with MEG. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.05.056. (Special issue on Mapping the Connectome) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5:11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Milchenko M, Marcus DS. Obscuring surface anatomy in volumetric imaging data. Neuroinformatics. 2013;11:65–75. doi: 10.1007/s12021-012-9160-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Loughead J, Ruparel K, Elliott MA, Hakonarson H, Gur RC, Gur RE. Impact of in-scanner head motion on multiple measures of functional connectivity: relevance for studies of neurodevelopment in youth. Neuroimage. 2012;60:623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Moeller S, Xu J, Auerbach EJ, Woolrich MW, Beckmann CF, Jenkinson M, Andersson J, Glasser MF, Van Essen DC, Feinberg DA, Yacoub ES, Ugurbil K. Temporally-independent functional modes of spontaneous brain activity. Proc Natl Acad Sci U S A. 2012;109:3131–3136. doi: 10.1073/pnas.1121329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos SN, Jbabdi S, Xu J, Andersson JL, Moeller S, Auerbach EJ, Glasser MF, Hernandez M, Sapiro G, Jenkinson M, Feinberg DA, Yacoub E, Lenglet C, Van Essen DC, Ugurbil K, Behrens TEJ. Advances in diffusion MRI acquisition and processing in the Human Connectome Project. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.05.057. (Special issue on Mapping the Connectome) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafyllou C, Hoge RD, Krueger G, Wiggins CJ, Potthast A, Wiggins GC, Wald LL. Comparison of physiological noise at 1.5 T, 3 T and 7 T and optimization of fMRI acquisition parameters. Neuroimage. 2005;26:243–250. doi: 10.1016/j.neuroimage.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Triantafyllou C, Polimeni JR, Wald LL. Physiological noise and signal-to-noise ratio in fMRI with multi-channel array coils. Neuroimage. 2011;55:597–606. doi: 10.1016/j.neuroimage.2010.11.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JA, Laird AR. The cognitive paradigm ontology: design and application. Neuroinformatics. 2012;10(1):57–66. doi: 10.1007/s12021-011-9126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugurbil K, et al. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.05.012. (Special issue on Mapping the Connectome) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith S, Barch D, Behrens TEJ, Yacoub E, Ugurbil K. The WU-Minn Human Connectome Project: an Overview. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.05.041. (Special issue on Mapping the Connectome) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Drury HA, Dickson J, Harwell J, Hanlon D, Anderson CH. An integrated software suite for surface-based analyses of cerebral cortex. J Am Med Inform Assoc. 2001;8:443–459. doi: 10.1136/jamia.2001.0080443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisskoff RM. Simple measurement of scanner stability for functional NMR imaging of activation in the brain. Magn Reson Med. 1996;36:643–645. doi: 10.1002/mrm.1910360422. [DOI] [PubMed] [Google Scholar]