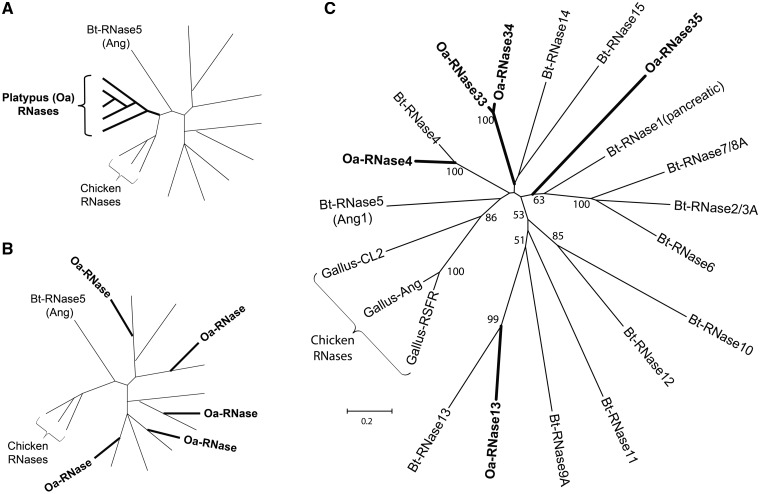
Fig. 2.—
Testing two hypotheses on the origin of the 13 ancient RNase gene lineages using a phylogenetic tree of five platypus (Oa), 13 cow (Bt), and three chicken (Gg) RNase genes. Two hypothetical trees (A and B) were made arbitrarily just to make a contrast between two hypotheses, and the real-data tree (C) was made using real sequences. The tree in panel (A) supports the hypothesis that the 13 ancient RNase gene lineages, represented by the 13 cow RNases, arose after the divergence between the platypus, whereas the tree in panel (B) supports the hypothesis that they arose before the divergence. The names and branches of the platypus RNases are bolded. In two hypothetical trees (A and B), branches without labels represent the cow RNases other than Bt-RNase5 (angiogenin), which is most closely related to nonmammalian RNases such as the chicken RNases. For the tree in panel (C), a total of 105 amino acid sites are used in tree making. We used the neighbor-joining methods with p distance and 1,000 bootstrap replications. The scale bar number indicates the number of amino acid substitutions per site.