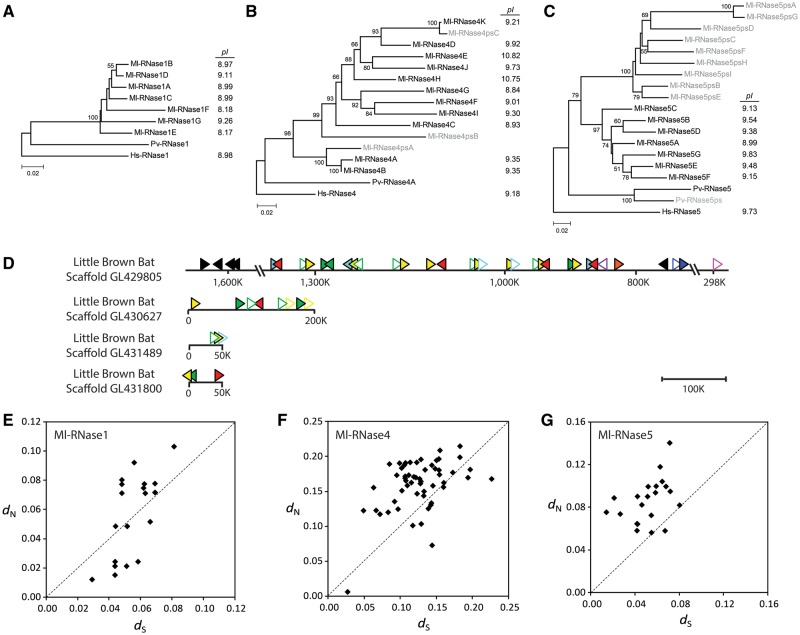
Fig. 5.—
Phylogenetic trees of the RNase1 (A), RNase4 (B), and RNase5 (C) genes of the littler brown bat (Ml) and the flying fox (Pv, Pteropus vampyrus). The human (Hs) genes were used to root the tree. The isoelectric points of the human and little brown bat proteins are shown next to the functional genes in three tree. (D) Scaffold maps of the little brown bat RNase genes drawn to scale. We followed the same labeling system as figure 1. Pairwise synonymous (dS) and nonsynonymous (dN) nucleotide distances between paralogous RNase1 (E), RNase4 (F), and RNase5 (G) genes in the little brown bat. The scale bar number indicates the number of nucleotide substitutions per site.