Abstract
The complex inner structures of concept names in the Foundational Model of Anatomy (FMA) remain an obstacle for further analyzing the ontology using lexical methods. A very common problem is the ambiguity lying in names with the sometimes multiple occurrences of the preposition “of.” In this paper, we propose an automatic method to help disambiguating FMA terms by leveraging the taxonomy and partonomy information. If a sub-phrase of a concept name also appears in its parents, it is likely to occur as a sub-tree in its parse tree, hence should be parsed as such. We classified all the concept names with a single occurrence of the preposition “of” by the appearances of their sub-phrases in the parent names using three test suites. Results show that more than 90% of them can be provided with useful information to assist their correct parsing.
Introduction
There exist several approaches for auditing biomedical ontologies [ 1 , 2 , 3 , 4 ]. Some of the approaches make use of lexical information in the terminology, requiring the understanding of the semantic structures in concept names [ 5 , 6 , 7 ]. For example, in our previous work [ 7 ], we provided an exhaustive, computationally scalable approach to uncover unidentified inconsistencies using inferred disjointness information in the Foundational Model of Anatomy (FMA) [ 8 ]. Antonymic modifiers were used therein to assume disjointness. However, for lengthier and more complex concept names in FMA (the longest one has 18 words), it remains a challenge to separate the “phrase stem” from the modifiers that contribute to disjointness, even with the recognition of antonymic modifiers. In such cases, Natural Language Process (NLP) tools, such as the well-known Stanford Parser, a probabilistic natural language parser for analyzing grammatical structure of sentences [ 9 ], may be employed to parse the concept names as a pre-processing step. However, when parsing the concept names in FMA using the Stanford Parser, only a small percentage of correct results were achieved on terms involving the preposition “ of ,” chiefly because of the ambiguity it introduces. Preposition induced ambiguity is an unsolved problem in noun phrase structure parsing [ 10 , 11 ].
For example, the concept name Left surface of heart has two parse trees due to different ways of scoping the modifier left ( Figure 1 ). For the parse tree on the left, the scope of left is surface of heart , while for the parse tree on the right, the scope of it is just surface .
Figure 1:

Two possible ways to parse the term “left surface of heart” as a noun phrase (NP). On the left, the scope of “left” is the noun phrase “surface of heart.” On the right, the scope of “left” is just “surface.”
When parsing FMA names using the Stanford Parser, we also found that it tended to use the style of tree on the right of Figure 1 for such terms. For example, the parse trees for the two concepts Mesothelial cell of parietal peritoneum and Left surface of heart both follow the style of the parse tree on the right in Figure 1 . However, while the first one is correct, the parse tree for Left surface of heart is not, because the scope of the modifier Left should be the entire string surface of heart instead of just surface . Typically, this type of ambiguity arises with the co-occurrence of the modifier (usually the first word) and the preposition “ of ” The scope of the modifier can be either the area before “ of ” or to the end of the entire string. If it is before “ of ,” the parse tree should follow the style of tree on the right of Figure 1 . Otherwise, it should follow the style of tree on the left of Figure 1 .
In this paper, we propose an automated method to help reducing ambiguity in FMA concept names by leveraging taxonomy and partonomy information. The corpus we choose is the set of all FMA concepts whose names contain a single occurrence of the preposition “ of ” and at least two words before “ of .” Among FMA’s entire 78,977 concepts, 61,877 of them contain at least one instance of preposition “ of ” and 35,174 of them contain a single occurrence. In the latter set, 14,153 terms have a single word before “ of ,” which must be a noun. Therefore, our study focuses on the remaining 21,021(about 27% of the total number of FMA concepts) concept names with at least two words before the preposition “ of ,” where the first words of those names are most likely to be modifiers.
For each concept name in this corpus, we create three sets of sub-phrases and use them as test suites to help determining the scope of the first modifier. The approach is quite simple: we go up one step to its parents and test for occurrences of sub-phrases. If a sub-phrase in the test suites also appears as sub-phrases in some of its parent names, this sub-phrase itself is assumed to be a sub-tree in the parse tree for the concept name, hence should be parsed as such. For good record keeping, we tag parent concepts either as an IS-A (taxonomy) parent or a Part-Of (partonomy) parent, according to the relationship type.
For the example Left surface of heart , we found this concept to be a subclass of Region of surface of heart and at the same time a part of Surface of heart . Our method suggests that surface of heart should form a subtree in the entire parse tree for Left surface of heart , implying that the correct parse tree should be the one on the left of Figure 1 . For Mesothelial cell of parietal peritoneum , it has parent Mesothelial cell and at the same time is a part of Mesothelium of parietal peritoneum . Our method suggests in this case that Mesothelial cell and parietal peritoneum should form sub-trees in the parse tree for Mesothelial cell of parietal peritoneum .
By reducing ambiguity, our method can more accurately parse FMA concept names and empower auditing methods that rely on lexical information.
1. Material
The FMA is both a theory of human anatomy and an ontology artifact [ 8 ]. For our analysis, we used the legacy version of the OWL [ 12 ] translation of FMA from the Open Biomedical Ontology (OBO) Foundry [ 13 ]. The concepts in FMA are organized into a concept hierarchy with more generic concepts modeled as parents of more specific concepts using the subClassOf property (e.g. Right Hand subClassOf Hand). This relationship is called the IS-A relationship. Another important relationship is the Part-Of relationship (e.g. Hand part_of Free upper limb). Usually, we use the taxonomy graph and the partonomy graph to represent the extracted IS-A and Part-Of relationships, respectively.
Our methodology leverages two technologies: one is the Semantic Web technology. The other one is the Stanford Parser [ 9 ]. In Semantic Web, RDF [ 14 ] is used as a universal data format to represent directed, labeled multi-graphs. The Web Ontology Language (OWL [ 12 ]) is a formal language for specifying the constraints of a particular domain, and is meant to govern the structure and meaning of the vocabulary used by RDF content. OWL ontologies can be serialized as RDF/XML graphs. SPARQL [ 15 ] is the query language for RDF, in the same way that SQL is the query language for relational databases.
The Stanford Parser [ 9 ] was trained on hand-parsed sentences to try to produce the most likely analysis of new sentences [ 16 ]. Even though it was strained on non-medical domain, it performs well in noun phrase identification in biomedical domain [ 17 ]. The parser provides Stanford typed grammatical dependencies as well as phrase structure trees [ 16 ]. The dependency structure output of Stanford Parser has been used to extract structured medical knowledge from biomedical domain [ 18 ].
2. Methods
In order to determine the scope of the modifier a 0 in the concept name S =( a 0 a 1 · · · a n of S 1 ), we create three test suites for it. Each one includes a set of sub-phrases. The first test suite is used to verify that the scope of a 0 is to the end of S . The second one is used to verify that the scope of a 0 ends before the preposition “ of .” The third test suite is used to check if a concept name has any ancestor in the affirmative results of the first two test suites. Below we give detailed information on the three test suites :
Test Suite 1 . Sub-phrases containing ( a n of S 1 ) but not ( a 0 ), i.e., sub-phrases from ( a 1 · · · a n of S 1 ) to ( a n of S 1 ). For example, Posterior papillary muscle of left ventricle has two sub-phrases in this set: papillary muscle of left ventricle and muscle of left ventricle . The principle for this case is simple: If a sub-phrase s in this test suite appears in both the IS-A parent and at least one Part-Of parent of S , it should be parsed as a sub-tree in the parse tree for S .
Test Suite 2 . The two sub-phrases ( a 0 a 1 · · · a n ) and ( of S 1 ). If the parents contain ( a 0 a 1 · · · a n ) but not ( a 0 a 1 · · · a n of ) (which indicates that ( a 0 a 1 · · · a n ) is independent of “ of ”), or, if the parents contain ( of S 1 ) but not ( a n of S 1 ) (which indicates that of is attached to S 1 ), the sub-phrase ( a 0 a 1 · · · a n ) is assumed to be a sub-tree in the parse tree for S . For example, Basivertebral foramen of thoracic vertebra is a subclass of Basivertebral foramen and at the same time a part of Body of thoracic vertebra , which indicates that Basivertebral foramen is an independent sub-phrase in the parse tree.
Test Suite 3 . The sub-phrase ( a 0 a 1 · · · a n of ). If the parents contain it, S is likely to have an ancestor such that the scope of the modifier in it can be determined by use of the first two test suites. For example, the concept Spinous process of tenth thoracic vertebra has an IS-A trace as follows: Spinous process of tenth thoracic vertebra → Spinous process of thoracic vertebra → Spinous process of vertebra → Process of vertebra . It is not until the last step that we can find the scope of spinous using the sub-phrase process of vertebra in the first test suite.
For each test suite, we classify the concept names into four categories along the possible combinations of appearance or non-appearance of the sub-phrases in its parent concepts. For a given concept name S and a test suite set T of it, the four categories are illustrated in Figure 2 : A. There exists s ∈ T such that s is a sub-phrase of the IS-A parent of S . At the same time, there also exists s′ ∈ T such that s′ appears as a sub-phrase in one of the Part-Of parents of S ; B. There exists s ∈ T such that s is a sub-phrase of the IS-A parent of S . However, no string in T appears as a sub-phrase in any of the Part-Of parents of S ; C. No string in T is a sub-phrase of the IS-A parent of S , but there exists s ∈ T such that s appears as a sub-phrase in one of the Part-Of parents of S ; D. No string in T is a sub-phrase of the IS-A parent of S . At the same time, no string in T appears as a sub-phrase in any of the Part-Of parents of S .
Figure 2:
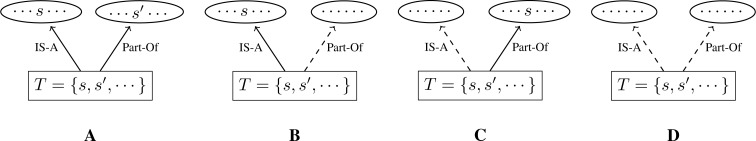
Four categories of child-parent relationships with respect to taxonomy (IS-A) and partonomy (Part-of). A : Some sub-phrase in T is contained in a parent term through IS-A, and some sub-phrase in T is also contained in a parent term through Part-Of. Both the sub-phrases and the parent terms may or may not be the same. B : Some sub-phrase in T is contained in a parent term through IS-A, but no sub-phrase in T is contained in any parent term through Part-Of. C : Some sub-phrase in T is contained in a parent term through Part-Of, but no sub-phrase in T is contained in any parent term through IS-A. D : Neither sub-phrase in T is contained in a parent term through Part-Of and nor sub-phrases in T is contained in any parent term through IS-A.
Note that for the second test suite, the criteria is stricter than others: As stated before, each occurrence of ( a 0 a 1 · · · a n ) should be accompanied by the nonoccurrence of ( a 0 a 1 · · · a n of ) and each occurrence of ( of S 1 ) should be accompanied by the nonoccurrence of ( a n of S 1 ).
Our method applies the three test suites one by one: The first round classifies all the concepts in the corpus with the first test suite. After that, for those concepts in Category D, the second round classifies them again with the second test suite. The four categories generated by the second round are renamed as Category DA, DB, DC and DD. At last, the third round applies the last test suite to Category DD, resulting in Category DDA, DDB, DDC and DDD. Therefore, after our analysis, each FMA concept term will fall precisely into one of the following ten categories: A, B, C, DA, DB, DC, DDA, DDB, DDC, DDD. Only terms in the category DDD (about 9%) do not exhibit any sub-phrases (determined by our test suites) in parent concepts.
For example, the term Spinous process of tenth thoracic vertebra has IS-A parent Spinous process of thoracic vertebra and two Part-Of parents: Tenth thoracic vertebra and Tenth thoracic vertebra arch . The first test suite contains only one sub-phrase process of tenth thoracic vertebra , which does not appear in any parents. So it is classified to Category D after the first round. The second test suite contains Spinous process and of tenth thoracic vertebra . Although the IS-A parent contains Spinous process , it also contains Spinous process of . As a result, the concept is classified to Category DD after the second round. The third test suite contains Spinous process of , which appears in the IS-A parent. At last, the concept is classified into Category DDB.
The following is the algorithm for the first round on an input concept name S =( a 0 a 1 a n of S 1 ) (each a i represents a word and S 1 represents the remaining sub-phrase). The algorithms for the second and third rounds maintain similar steps.
Extract the sub-phrase s 1 = ( a 1 a n of S 1 ).
Make a SPARQL Query to get the IS-A parent of S in the Virtuoso RDF store, if s 1 appears as a sub-phrase of the parent, set the variable mark_isa=true; else set the variable mark_isa=false.
Make a SPARQL Query to get all the Part-Of parents of S in the Virtuoso RDF store, if s 1 appears as a sub-phrase in one of them, set the variable mark_partof=true; else set the variable mark_partof=false.
If mark_isa=true and mark_partof=true, classify S into Category A, stop algorithm; If not, go to step 5.
If it is the sub-phrase s n = ( a n of S 1 ), classify S into the other three categories according to the values of mark_isa and mark_partof; If not, extract the next sub-phrase, then repeat the procedure from step 2 to step 4 for it.
3. Results
The number and percentage distribution of the ten categories, A, B, C, DA, DB, DC, DDA, DDB, DDC, DDD, are shown in Table 1 . The percentages are based on the total number of FMA concepts in the corpus(21,021).
Table 1:
Total counts and percentages of FMA terms into ten categories. Data on categories A, B, C appear in rows two, three, and four. Category D is broken into four columns. The first three columns provide data for categories DA, DB and DC. The fourth column provides the number for category DD, with 12709 terms representing 60.46% of total terms, which are further broken into DDA (245; 1.16%), DDB (10547; 50.17%), DDC (14; 0.06%) and DDD (1903; 9.05%).
| Count and Percentage Distribution | |||||||
|---|---|---|---|---|---|---|---|
| A | 321 (1.52%) | ||||||
| B | 3025 (14.39%) | ||||||
| C | 1044 (4.97%) | ||||||
| D | 16631 (79.11%) | ||||||
| DA | DB | DC | DD | ||||
| 396 (1.88%) | 2217 (10.55%) | 1309 (6.22%) | 12709 (60.46%) | ||||
| DDA | DDB | DDC | DDD | ||||
| 245 (1.17%) | 10547 (50.17%) | 14 (0.07%) | 1903 (9.05%) | ||||
The fact that half of the concept names in the entire corpus belong to DDB demonstrates that for those concept names, although the immediate parents cannot provide helpful information for parsing them, if we go up several steps to the ancestors, the chances are good to get affirmative information. In conclusion, except for the 1903 concept names in DDD, 19,118 (90.95%) concept names can be provided with information to assist their correct parsing.
4. Discussion and Conclusions
The amount of information each category can provide to help determine the scope of the modifier varies. Although Category A can provide decisive information, the requirement is also very strict. Concepts that do not have any Part-Of parent can only be classified into Category B or D. So we should also use Category B and Category C to help chunking the concept names. Since the IS-A relationship often leads to more abstract concepts, we conjecture that the Part-Of relationship may be more reliable, i.e., Category C may provide more information than Category B. For the 12,709 concept names in Category DD, the first two test suites both failed to provide useful information to determine the scope of the modifier. This is mainly caused by a limitation of our algorithm: only lexical information of the immediate upper neighborhood is used.
Our work has a couple of limitations as it stands. One is that only terms containing a single occurrence of “ of ” are considered in this paper. Future work would extend the method to multiple occurrences of the preposition “ of .” The second is that some terms may require several steps up in the hierarchy to obtain useful information, which is evident from the fact that more than 50% of the concepts in the corpus belong to Category DDB (see Table 1 ). We believe that if we expand the search pool from parents to ancestors or the entire set of FMA concept names, then the number of instances in those categories with affirmative information will increase, although the computational cost will also increase.
In [ 19 ], the authors also used substring relations to analyze term names in Gene Ontology. Different from our paper, their focus was on the relations between terms instead of the inner structure within each term name.
In conclusion, our work is an initiative step towards the utilization of an un-tapped information source to not only help improve the accuracy of lexical-based auditing methods, but also guide the initial phase for the creation of long and complex concept names in terms of proving information on how they will be parsed.
Acknowledgments
This publication was made possible by the Clinical and Translational Science Collaborative (CTSC) of Cleveland, UL1TR000439, from the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health and NIH roadmap for Medical Research and supplement UL1RR024989-05S from the National Center for Research Resources of NIH.
References
- 1. Bodenreider O . Quality Assurance in Biomedical Terminologies and Ontologies . Bethesda : Lister Hill National Center for Biomedical Communications, National Library of Medicine ; 2010 . [Google Scholar]
- 2. Geller J , Perl Y , Halper M , Cornet R . Guest Editorial: Special Issue on Auditing of Terminologies . Journal of Biomedical Informatics . 2009 June ; 42 ( 3 ): 407 – 411 . doi: 10.1016/j.jbi.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 3. Zhu X , Fan JW , Baorto DM , Weng C , Cimino JJ . A Review of Auditing Methods Applied to the Content of Controlled Biomedical Terminologies . J Biomed Inform . 2009 ; 42 ( 3 ): 413 – 25 . doi: 10.1016/j.jbi.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang GQ , Bodenreider O . Large-scale, Exhaustive Lattice-based Structural Auditing of SNOMED CT . AMIA Annu Symp Proc . 2010 ; 2010 : 922926 . [PMC free article] [PubMed] [Google Scholar]
- 5. Bodenreider O , Burgun A , Rindflesch TC . Assessing the consistency of a biomedical terminology through lexical knowledge . International Journal of Medical Informatics . 2002 ; 67 ( 1–3 ): 85 – 95 . doi: 10.1016/s1386-5056(02)00051-5. [DOI] [PubMed] [Google Scholar]
- 6. Zhang S , Bodenreider O . Aligning representations of anatomy using lexical and structural methods . AMIA Annu Symp Proc . 2003 : 753 – 757 . [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang GQ , Luo L , Ogbuji C , Joslyn C , Mejino J , Sahoo SS . An Analysis of Multi-type Relational Interactions in FMA Using Graph Motifs . AMIA Annu Symp Proc . 2012 [PMC free article] [PubMed] [Google Scholar]
- 8. Rosse C , Mejino JLV . A Reference Ontology for Bioinformatics: The Foundational Model of Anatomy . Journal of Biomedical Informatics . 2003 ; 36 : 478 – 500 . doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 9. Klein D , Manning CD . Accurate Unlexicalized Parsing . Proceedings of the 41st Meeting of the Association for Computational Linguistics . 2003 : 423 – 430 . [Google Scholar]
- 10. Vadas D , Curran JR . Adding Noun Phrase Structure to the Penn Treebank . ACL ; 2007 . [Google Scholar]
- 11. Vadas D , Curran JR . Parsing Noun Phrases in the Penn Treebank . Computational Linguistics . 2011 ; 37 ( 4 ): 753 – 809 . [Google Scholar]
- 12. Hitzler P , Krotzsch M , Parsia B , Patel-Schneider PF , Rudolph S . OWL 2 Web Ontology Language Primer: W3 C Recommendation . 2009 .
- 13. OBO FOUNDRY http://www.obofoundry.org/
- 14. RDF http://www.w3.org/RDF/
- 15. SPARQL http://www.w3.org/TR/rdf-sparql-query/
- 16. Marneffe MC , MacCartney B , Manning CD . Generating Typed Dependency Parses from Phrase Structure Parses . LREC ; 2006 . [Google Scholar]
- 17. Xu R , Supekar K , Morgan A , Das A , Garber AM . Unsupervised Method for Automatic Construction of a Disease Dictionary from a Large Free Text Collection . Annual American Medical Informatics Association Symposium . 2008 : 820 – 824 . [PMC free article] [PubMed] [Google Scholar]
- 18. Xu R , Das A , Garber AM . Unsupervised Method for Extracting Machine Understandable Medical Knowledge from a Large Free Text Collection . Annual American Medical Informatics Association Symposium . 2009 : 709 – 713 . [PMC free article] [PubMed] [Google Scholar]
- 19. Ogren PV , Cohen KB . The compositional structure of Gene Ontology terms . Pac Symp Biocomput . 2004 : 214 – 25 . doi: 10.1142/9789812704856_0021. etc . [DOI] [PMC free article] [PubMed] [Google Scholar]


