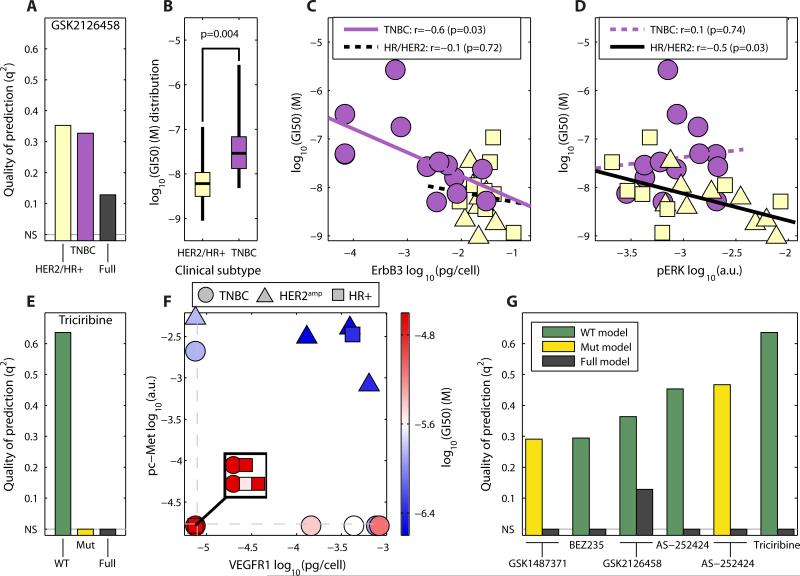
Fig. 5. Inclusion of clinical and genomic information can improve predictions of drug response.
(A) Quality of the prediction for models stratified according to subtype (TNBC versus HR+/HER2amp) for the pan-PI3K inhibitor GSK2126458. Predictions by subset are shown according to a model built on just the indicated subset of cell lines (yellow and purple bars) or all of them (black bar). (B) Distribution of GI50 values for GSK2126458 by subtype. (C, D), Correlation of ErbB3 (C) and pERK (D) with GSK2126458 GI50 values for each of the three cancer subtypes. A dashed line shows a non-significant correlation. (E) Quality of the prediction of models stratified according to mutational status of PIK3CA and PTEN for triciribine. Full, all cell lines; WT cell lines wild-type for both PIK3CA and PTEN; Mut, cell lines with mutations in either or both of PIK3CA and PTEN. Non-significant predictions (NS) are set to zero. (F) Projection of the abundance of pc-MET and VEGFR1 overlaid with the triciribine GI50 values only for cell lines wild-type for PIK3CA and PTEN (red, resistant; blue, sensitive). Dashed lines show the detection threshold for each measurement. (G) Quality of the prediction of models stratified according to mutational status of PIK3CA and PTEN (WT versus Mut) for drugs targeting the PI3K/Akt pathway. Only those drugs for which stratification improves prediction are shown.