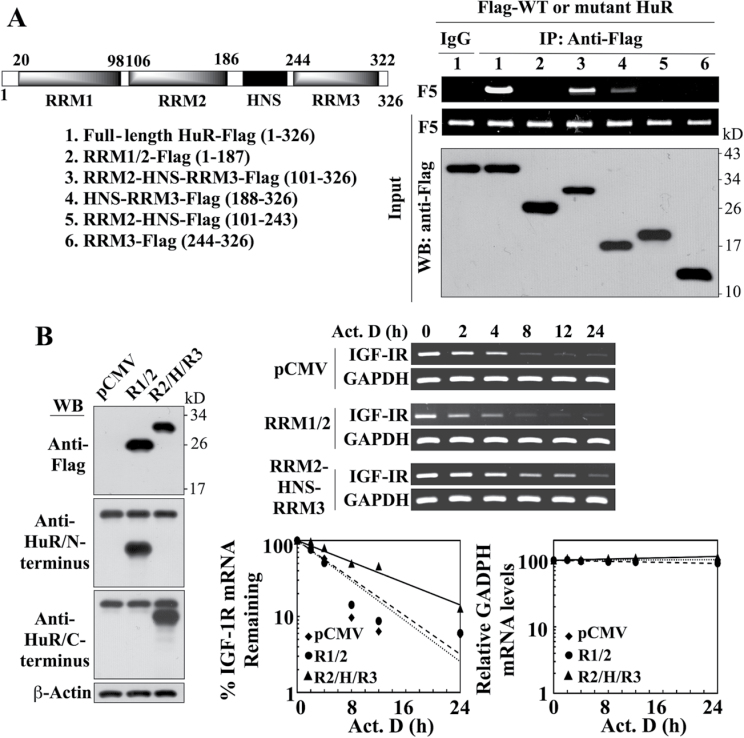
Fig. 8.
Evidence that the portion of HuR encompassing the RRM2-HNS-RRM3 motifs is involved in mediating HuR-facilitated IGF-IR mRNA stabilization. (A) Left, schematic representation of the structures of Flag-tagged wild-type and various truncated mutant forms of HuR. Right, immunoprecipitation analysis of the interaction of wild-type HuR versus truncated HuR mutants with the IGF-IR 3′UTR-F5. LNCaP cells were transiently cotransfected with Flag-tagged wild-type HuR or individual truncated mutants for 48h, followed by immunoprecipitation (IP) with anti-Flag- or anti-IgG agarose conjugates. The abundance of IGF-IR 3′UTR-F5 associated with the immune complexes was analyzed by RT–PCR. (B) Left, western blot analysis depicting the abundance of ectopically expressed Flag-tagged RRM1-RRM2 (R1/2) and RRM2-HNS-RRM3 (R2/H/R3) HuR mutants in transiently transfected LNCaP cells. Because commercial antibodies recognize only either the N- or C-terminus of HuR, two different mAbs were used to immunostain R1/2 and R2/H/R3, respectively. Right upper, RT–PCR analysis of the stability of IGF-IR mRNA in LNCaP cells transiently transfected with the RRM1-RRM2 versus the RRM2-HNS-RRM3 HuR mutant or pCMV control in the presence of 10 µM actinomycin D (Act. D) in 10% FBS-supplemented RPMI 1640 medium. Right lower, densitometric analysis of the abundance of IGF-IR versus GAPDH mRNA remaining in cells at the indicated time intervals per the RT–PCR analysis (n = 3). Amounts of mRNA are expressed as a percentage of that present at the 0h time point on a log scale. All experiments were performed three times with similar results.