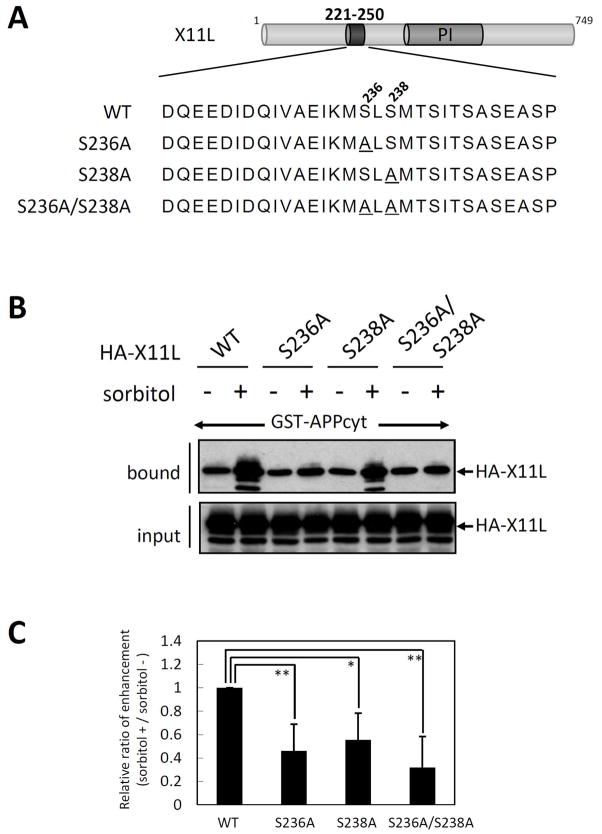
Figure 7. Role of Ser236 and Ser238 in X11L221-250 in the enhanced association with APPcyt.
(A) Sequences of amino acid substitution mutants at Ser236 and/or Ser238. Alanyl residues substituted for Ser236 (S236A) and Ser238 (S238A) are underlined. WT indicates the wild-type X11L. PI indicates the PI/PTB domain.
(B) Association of APPcyt with X11L mutants derived from sorbitol treated cells. HEK293 cells (~2 × 105) were transiently transfected with 0.5 μg of pcDNA3-HA-X11L (WT), pcDNA3-HA-X11L S236A (S236A), pcDNA3-HA-X11L S238A (S238A), or pcDNA3-HA-X11L S236A/S238A (S236A/S238A), and treated with (+) or without (−) 0.5 M sorbitol for 45 min. The cell lysates were incubated with GST-APPcyt, and HA-X11L proteins bound to APPcyt (bound) and the lysates (input) were analyzed by immunoblotting with anti-HA 12CA5 antibody.
(C) Quantification of X11L proteins bound to APPcyt. X11L proteins bound to APPcyt were quantified with the VersaDoc imaging system (Bio-Rad). The data are presented as the ratio (sorbitol (+)/sorbitol (−)) relative to the level of wild type X11L-binding (WT, 1.0). Results represent means ± s.d. for n=4 experiments. *, p<0.05, **, p<0.01 by Student’s t-test.