Figure 2. Compressed sensing (CS) selection of PGT-123 epitope residues.
Results of the application of the compressed sensing classification algorithm to the neutralization activity of bnMAb PGT-123 against a panel of 141 HIV-1 pseudoviruses (cf. Table S1). In each panel, the abscissa indicates the number of non-zero elements in the 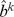 signal vector computed by the LASSO algorithm, and therefore the number of residues incorporated into the regularized least squares fit of the neutralization data (Eqn. 3). For clarity of viewing, plots are terminated at the 100-component model. As indicated by the arrows, knees in the (a) mean squared error (MSE) over the complete data set and (b) leave-one-out cross-validation mean squared error (LOOCV-MSE) curves were identified using the L method at 11 and 9 residues, respectively [77]. The mean of these values motivated the selection of the ten residues constituting this model: I323, H330, N332, N334, S334, S612, N671, Q740, V815, and V843 (c.f. Table 1).
signal vector computed by the LASSO algorithm, and therefore the number of residues incorporated into the regularized least squares fit of the neutralization data (Eqn. 3). For clarity of viewing, plots are terminated at the 100-component model. As indicated by the arrows, knees in the (a) mean squared error (MSE) over the complete data set and (b) leave-one-out cross-validation mean squared error (LOOCV-MSE) curves were identified using the L method at 11 and 9 residues, respectively [77]. The mean of these values motivated the selection of the ten residues constituting this model: I323, H330, N332, N334, S334, S612, N671, Q740, V815, and V843 (c.f. Table 1).

