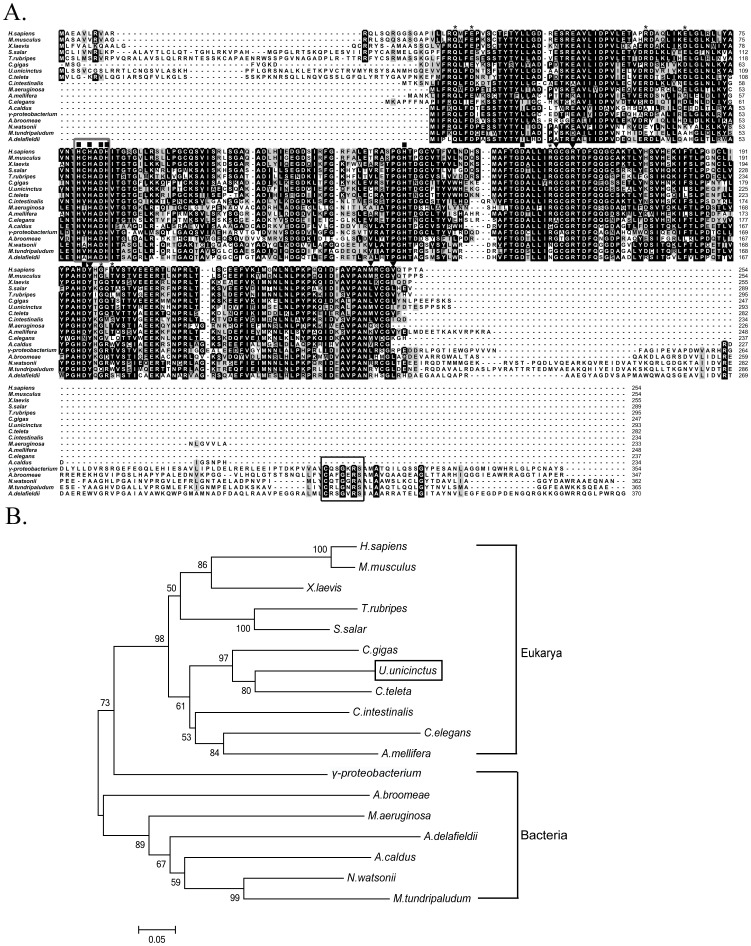
Figure 6. Multiple sequence alignment (A) and phylogenetic relationships (B) among the SDO sequences from different species.
Identical and similar residues are highlighted in black and gray, respectively. Conserved relevant metal binding sites, GSH binding sites and dimer formation residues are indicated by ▪, ▾ and *, respectively. The β-lactamase signature motif and rhodanese active-site loop are marked with a black box and grey box, respectively. GenBank accession numbers: Afipia broomeae (ZP_11429388.1), Acidithiobacillus caldus (YP_004749948.1), Acidovorax delafieldii (ZP_04761469.1), Apis mellifera (XP_393510.1), Caenorhabditis elegans (NP_501684.1), Capitella teleta (JGI Genome), Ciona intestinalis (XP_002128021.1), Crassostrea gigas (EKC28487.1), Homo sapiens (NP_055112.2), Methylobacter tundripaludum (ZP_08782165.1), Microcystis aeruginosa (ZP_18834377.1), Mus musculus (NP_075643.1), Nitrosococcus watsonii (YP_003760989.1), γ-proteobacterium HTCC2148 (ZP_05095460), Salmo salar (ACI68458.1), Takifugu rubripes (XP_003977175.1), Urechis unicinctus (AEV92813.1), Xenopus laevis (NP_001079404.1).