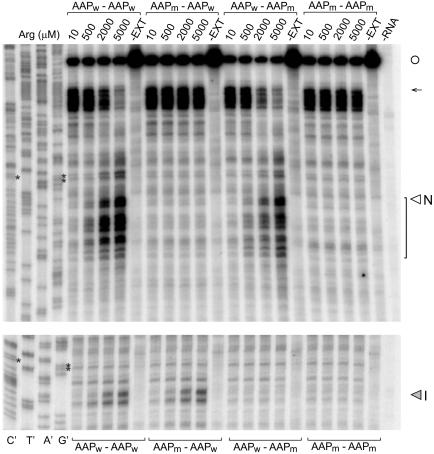
Fig. 3.
Toeprint analysis of ribosome stalling at AAP domains in Met9-AAP-globin-AAP-LUC mRNA. Separate gels and primers were used for analyses of stalling at the N-terminal (Upper) and internal (Lower) AAP domains for optimal resolution. N-terminal and internal AAP domains are indicated as wild-type (AAPw)or mutated (AAPm) above or below the corresponding lanes. Transcripts were translated in 20-μl reaction mixtures containing 10, 500, 2,000, or 5,000 μM Arg as indicated and 10 μM of the other amino acids and analyzed as described (13, 25). (Left) Sequencing reactions for the Met9-AAPw-globin-AAPw-LUC template. The sequence can be directly read 5′ to 3′ from top to bottom. Controls: Products obtained from primer extension of RNA (18 ng) in the absence of extract (–EXT) and from an extract not programmed with RNA (–RNA). Primers FP94 and FP93 (Fig. 5A) were used for the experiments shown (Upper and Lower, respectively). The open circle indicates the mRNA 5′ end; the arrow indicates the position of ribosomes at the first Met (start) codon. Translation of the nine contiguous Met codons was slow, as evidenced by the toeprints of ribosomes in this region. Asterisks mark each AAP's final GCG codons, which lie ≈16 nt upstream of the toeprints corresponding to the stall sites associated with the production of polypeptide intermediates N and I (indicated); toeprints that represent translational stalling after the Met9-AAP coding region are indicated with a bracket.