| Step 1. |
Identify all pairs of positions i,j in the sequence alignment where at least 20% of the residue pairs have change, volume, and hydrophobicity that lie in the same 3D property bin φpqr. |
| Step 2. |
Evaluate mutual information 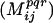 score for all pairs of positions i,j denoted as conserved for the corresponding bin φpqr. score for all pairs of positions i,j denoted as conserved for the corresponding bin φpqr. |
| Step 3. |
Perform bootstrap replicate analysis and select positions i,j that meet the P value cutoff of 5 × 10-3. |
| Step 4. |
Investigate the selected residue positions in the hybrids for clashes based on the following criteria: |
|
|
| Step 5. |
Hybrids are given a score equal to the number of clashes identified in step 4. |
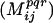 score for all pairs of positions i,j denoted as conserved for the corresponding bin φpqr.
score for all pairs of positions i,j denoted as conserved for the corresponding bin φpqr. 
