Figure 1.
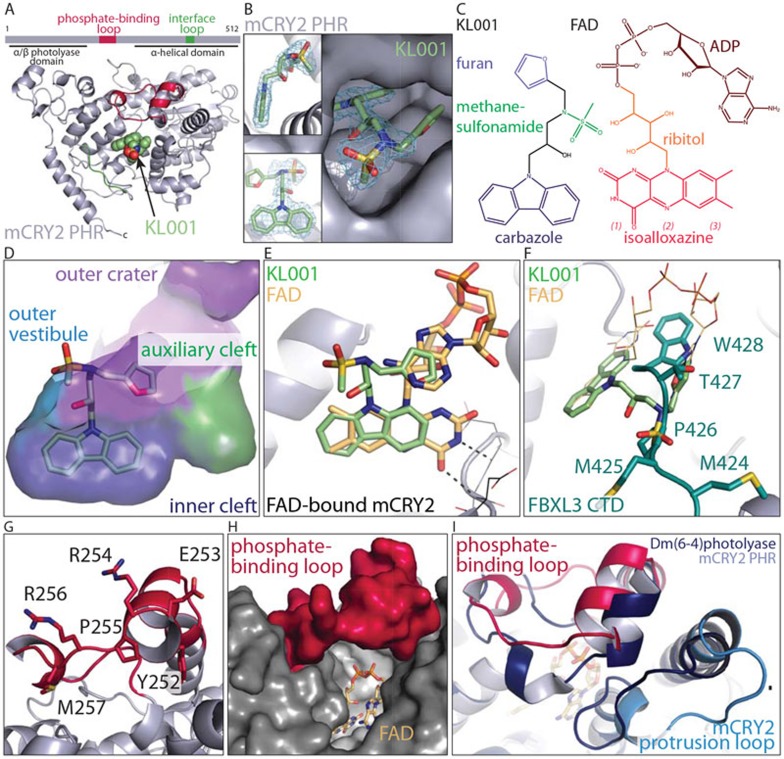
Structure of murine CRY2 in complex with KL001.(A) Overall structure of KL001-bound mCRY2 PHR domain. (B) Three close-up views of KL001 bound to CRY2 with positive Fo-Fc electron density contoured at 3.5 σ and calculated before the compound was built. (C) FAD and KL001 with their functional groups labeled. Subdivisions of the isoalloxazine ring: (1) pyrimidine, (2) pyrazine, (3) xylene. (D) Docking of the KL001 carbazole ring at the inner cleft (blue) of the FAD-binding pocket with the nearby auxiliary cleft (green), the outer crater (purple) and part of the outer vestibule shown. (E) A comparison of KL001 and FAD bound to CRY2. FAD is modeled onto the KL001-bound CRY2 (ribbon diagram) by CRY2 superposition. Two hydrogen bonds formed between the backbone of FAD-bound CRY2 (line representation) and the pyrimidine moiety of the FAD isoalloxazine ring are shown. (F) A different view of CRY2-bound KL001 in comparison with FAD (line representation) and the C-terminal tail of FBXL3 (teal). (G) Conformation of the ordered phosphate-binding loop of CRY2 with highly conserved but solvent exposed residues. (H) A model of FAD-bound CRY2 with an ordered phosphate-binding loop showing an open cofactor pocket. (I) A structural comparison between KL001-bound murine CRY2 (grey) and Drosophila(6-4)photolyase (dark blue) showing the phosphate-binding loop (pink in CRY2) and its nearby protrusion loop (light blue in CRY2). FAD is modeled in to replace KL001.