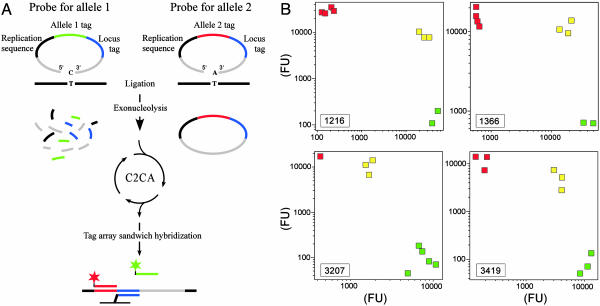
Fig. 3.
PCR-independent multiplexed genotyping. (A) Allele-specific padlock probes were constructed with 20 target-complementary nucleotides at both the 5′ and 3′ ends (gray) and with the allele-specific nucleotide positioned at the 3′ end. Target-specific ends were connected by the following three segments: a replication sequence common for all probes (black), either of two allele tag sequences (red or green), and finally a locus tag sequence (blue). Probe arms correctly hybridized to a target sequence (black line) are efficiently joined by a DNA ligase. A subsequent exonuclease treatment degrades unreacted probes while preserving circularized ones. Circularized probes serve as templates in a replication reaction primed by the general replication oligonucleotide (RO). The monomerized third-generation RCR products were applied to a tag microarray along with TAMRA- or FITC-labeled (red or green star) allele-tag specific oligonucleotides in a sandwich hybridization. (B) Results from a multiplexed genotyping experiment of nine different genomic DNA samples using a pool of 26 padlock probes. Signals from the different allele-specific amplification products were plotted in log–log diagrams for 4 of the 13 loci. The genotypes of the samples were known from previous studies. Green squares represent individuals homozygous for allele 1, red squares represent individuals homozygous for allele 2, and yellow squares represent heterozygous individuals. Locus identities are identified in the lower left corners.