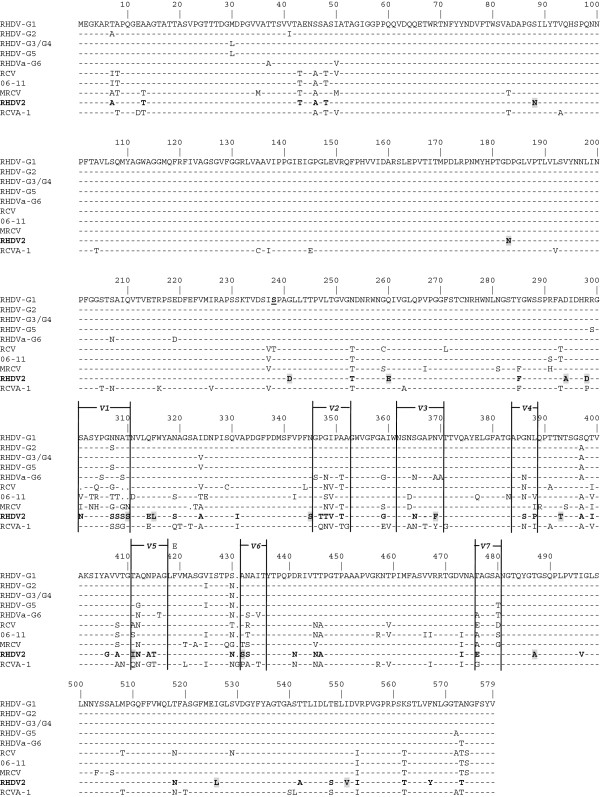
Figure 1.
Consensus amino acid sequence alignment of the VP60 gene from RHDV2 and other lagoviruses. The aligned lagovirus sequences correspond to the consensus sequences of i) pathogenic RHDV and RHDVa sequences (47 sequences) belonging to the different genetic groups (G1 to G6) identified by Le Gall-Reculé et al. [16], ii) RCV-A1 sequences (36 sequences). Three other rabbit lagovirus sequences were aligned and correspond to the non-pathogenic RCV (X96868), the non-pathogenic 06–11 (AM268419) and the moderately pathogenic MRCV (GQ166866) strains. Residues differing from the first sequence are shown and a dot corresponds to a deletion. The hypervariable regions V1 to V7 according to Wang et al. [43] are indicated at the top of the alignment. The amino acid S238 (in bold and underlined in the first sequence) corresponds to the beginning of the protrusion domain of the VP60 protein [43]. The RHDV2 mutated amino acids shared by no other rabbit lagovirus are highlighted in grey.