Figure 3.
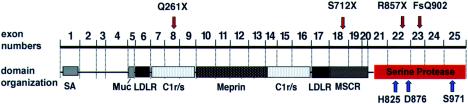
Position of mutations (red arrows), in relation to proenteropeptidase exon organization, domains, and amino acid residues forming the active site of the serine protease domain (H825, D876, and S971 [blue arrows]). All four mutations identified are null mutations predicting the absence of a correctly formed active site. The previously described modular structure of proenteropeptidase domains, based on primary-structure comparison, correlates with exon boundaries. SA = signal/anchor sequence; LDLR = LDL receptor–like domain; Muc = mucin-domain; Meprin = meprin-like domain; C1r/s = complement component C1r-like domain; MSCR = macrophage scavenger receptor–like domain.
