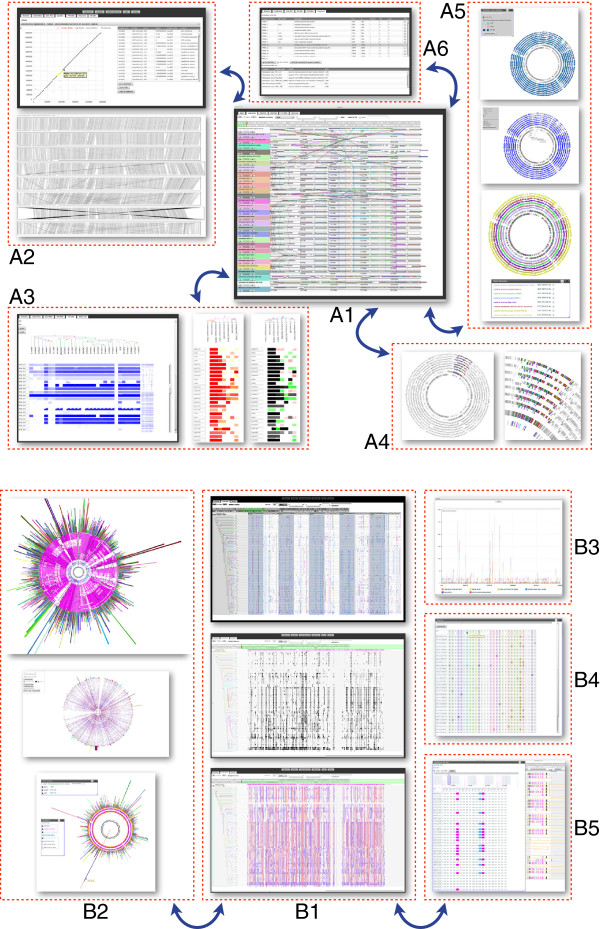
Figure 1.
Multiple connected views of the SynTView graphical user interface. Panel A shows several representations of genome syntenic organisations and protein similarity relationships. A1: Central view showing a genome browser with dynamic links between one reference genome (top) and several synteny-aligned related genomes; A2: Dot plot (with a connected gene table on the right-hand side) and line plot representations; A3: Phylogenetic profiles representing either the whole reference genome (left-hand side) or a defined gene set (right-hand side); the colour and shape of the trapezoidal box represent the sequence similarity and protein coverage, respectively; A4: In contrast to the central local view, where absolute coordinates of aligned genomes do not coincide, this global circular view indicates whether the conserved syntenic region is localised identically in complete genomes; A5: Several kinds of circular representations where the outer circle colours represent either sequence similarity heat maps (top) or genome categories (bottom); A6: Gene table with selection widgets and BDBH display. Panel B illustrates the exploration of polymorphism information. B1: Three linear views of phylogenetically ordered genomes showing information for SNPs at various scales and in various representation modes: individual SNPs coloured according to their location in relation to genes (top), SNP clusters (middle) and SNP heat maps (bottom); B2: Several circular representations of SNP distributions in n genomes compared to one reference genome (centre), the outside bars indicate the cumulated number of SNPs per gene; B3: Histogram of metadata-clustered SNPs enabling a differential analysis by groups of strains; B4: Artificial sequence reconstituted with polymorphic positions; B5: Gene browser representing the effect of mutated positions on the nucleotide or protein sequence. The blue arrows represent the dynamic integration between the views.