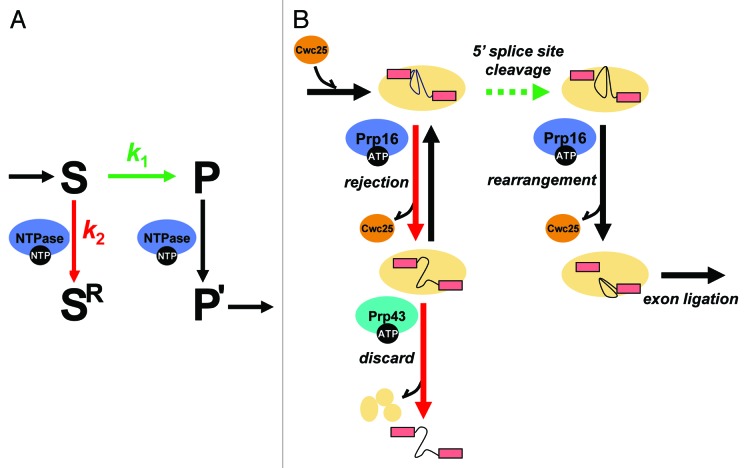
Figure 1. An ATP-dependent framework for kinetic proofreading by DEAD/H-box ATPases. (A) A general framework for kinetic proofreading by NTPases. In the kinetic proofreading scheme, an NTPase increases the specificity of a step (S → P, shown in green) by competing with it (S → SR, shown in red). In this scheme, k1 represents the rate of proceeding on-pathway, such as through 5′ splice site cleavage during pre-mRNA splicing, while k2 represents the rate of rejecting a substrate. Specificity of the step under inspection (S → P) is enhanced when the ratio of k1/k2 is higher for an optimal substrate than a suboptimal substrate. In this scheme, a proofreading NTPase not only antagonizes a suboptimal substrate (S → SR) but also promotes an optimal substrate (P → P’, shown in black), if the NTPase functions after, rather than before, the step under inspection. (B) A molecular framework by which the DEAH-box ATPase Prp16 establishes specificity during 5′ splice site cleavage. For an optimal substrate, Prp16 promotes splicing by enabling rearrangements after 5′ splice site cleavage that dissociate Cwc25, allowing repositioning of the reaction intermediates for exon ligation. For a suboptimal substrate, Prp16 antagonizes splicing, rejecting the substrate, by enabling rearrangements before 5′ splice site cleavage that similarly dissociate Cwc25 but also prime the spliceosome for discard of the substrate by the DEAH-box ATPase Prp43. In the kinetic proofreading model, specificity of 5′ splice site cleavage can be established if k5′ splice site cleavage (optimal substrate) > k5′ splice site cleavge (suboptimal substrate) and/or if krejection(suboptimal substrate) > krejection(optimal substrate). Our data provided evidence for the former case and that this proofreading NTPase functions, minimally, as a timer, setting a standard for the rate of 5′ splice site cleavage.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.