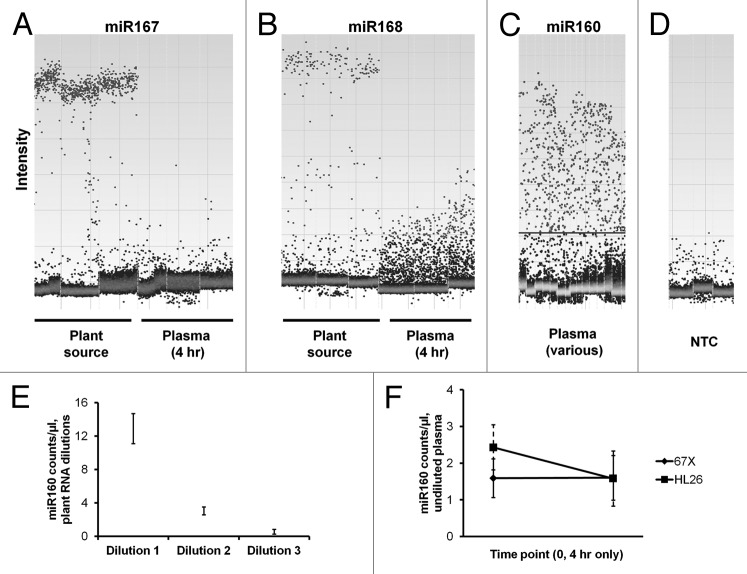
Figure 4. Droplet digital PCR indicates specific amplification of plant miRNA from plant source but not from pre- or postprandial animal plasma. (A and B) Intensity plots of DX100 QuantaSoft results for miR167 and miR168: for RNA from the dietary plant material (left), positive droplets clustered at high and relatively consistent intensity (specific amplification). Droplets from four hour postprandial plasma samples (right) had low and varying intensity, especially for miR168, consistent with inefficient or spurious amplification. Triplicate reactions are shown. Note that the intensity scale is not necessarily equivalent between different plots. (C) miR160 showed the highest level of amplification from plasma for miRNAs we examined by RT-qPCR (see Fig. 2), but by ddPCR with pre- and post-prandial plasma samples, there was a wide range of droplet intensities. From plant material, droplet intensities clustered consistently as for the miRNAs in (A and B) (not shown). The horizontal line is an example of where a threshold might be drawn. (D) Low-level non-specific amplification for plant miRs166, 167 and 168 from one no-template control reaction each (water). (E) Copy number counts and confidence interval minimum and maximum for miR160 per microliter of reaction volume over a 1:4 dilution series of material corresponding to cDNA from input into a reverse transcription reaction that corresponded to RNA obtained from 6.25 nl (dilution 1), 1.6 nl (dilution 2) and 400 pl (dilution 3) of the “Silk” plant-based substance. (F) Counts and confidence intervals for undiluted material from plasma RNA isolations for the pre-intake and 4 h postprandial time points for miR160 for subjects 67X and HL26.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.