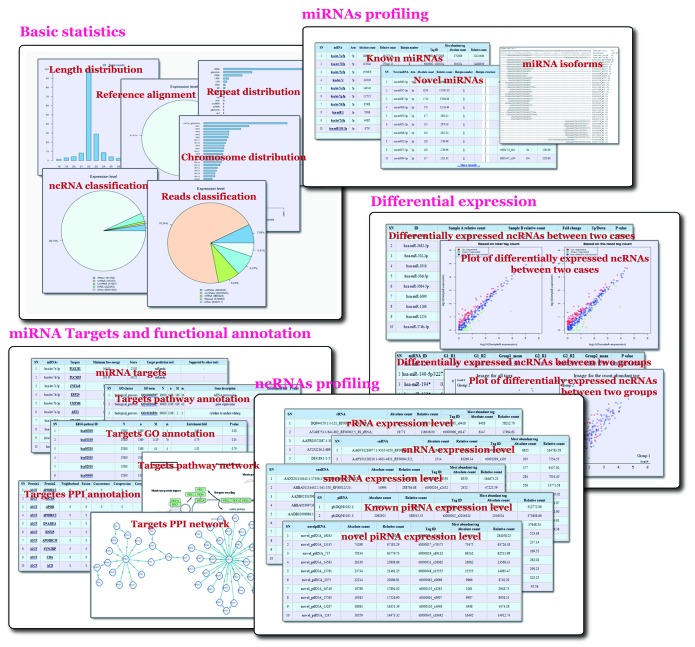
Figure 1. Output screenshots of mirTools 2.0. The output includes: (1) basic statistics, such as length distribution, percentage of reads aligned to the reference genome, chromosome distribution, functional categories of reads and ncRNA distribution; (2) known miRNA, putative novel miRNA, miRNA isoforms and modification; (3) miRNA-targeted genes and functional annotation based on GO, the KEGG pathway and the PPI network; (4) expression information of other types of ncRNA, such as rRNA, tRNA, snRNA, snoRNA and piRNA; and (5) differentially expressed ncRNAs between two cases, two experimental groups or among multiple samples.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.