Abstract
MicroRNAs (miRNAs) and other small non-coding RNAs (sncRNAs) are post-transcriptional regulators of gene expression, playing key roles in neuronal development, plasticity, and disease. Transcriptome deregulation caused by miRNA dysfunction has been associated to neurodegenerative diseases. Parkinson disease (PD) is the second most common neurodegenerative disease showing deregulation of the coding and small non-coding transcriptome. On profiling sncRNA in PD brain areas differently affected, we found that upregulation of a small vault RNA (svtRNA2-1a) is widespread in PD brains, occurring early in the course of the disease (at pre-motor stages). SvtRNA2-1a biogenesis was dependent on Dicer activity on its precursor (vtRNA2-1) but independent of Drosha endonuclease, unlike the canonical miRNAs. Although endogenous svtRNA2-1a was enriched in Ago-2 immunoprecipitates in differentiated SH-SY5Y neuronal cells, overexpression of svtRNA2-1a induced subtle transcriptomic changes, suggesting that gene expression regulation may involve other mechanisms than mRNA decay only. Function enrichment analysis of the genes deregulated by svtRNA2-1a overexpression or svtRNA2-1a predicted targets identified pathways related to nervous system development and cell type specification. The expression pattern of svtRNA2-1a during development and aging of the human brain and the detrimental consequences of a svtRNA2-1a mimic overexpression in neuronal cells further indicate that low svtRNA2-1a levels may be important for the maintenance of neurons. Our results suggest that early svtRNA2-1a upregulation in PD may contribute to perturbations of gene expression networks, underlying metabolic impairment and cell dysfunction. A better understanding of the pathways regulated by svtRNA2-a, and also the mechanisms regulating its expression should facilitate the identification of new targets for therapeutic approaches in PD.
Keywords: lincRNAs, microarrays, neuronal dysfunction, parkinson disease, sncRNA, vault RNA
Introduction
Parkinson disease (PD) is a progressive neurological disorder characterized by motor symptoms, such as tremor at rest, bradikynesia and postural instability as well as non-motor features, which include autonomic dysfunction, cognitive and neurobehavioral abnormalities as well as sleep and sensory disorders. The main pathological hallmark of PD is the loss of dopaminergic neurons in the substantia nigra (SN), in association with the presence of cytoplasmic protein inclusions named Lewy bodies (LB) and Lewy neurites (LN). Cell death in the SN, gives rise to the motor symptoms, which typically appear when there is a loss of about 50% of total SN neurons in comparison to age-matched controls, with a consequent depletion of 80% of the striatal dopamine.1-3 However, neuronal loss and protein inclusion pathology is not only restricted to dopaminergic neurons in the SN, but also occurs in the striatum and brain cortex, and further affects other cell types.4 The intracerebral formation of LBs and LNs has been proposed to begin at defined induction sites and advance in a topographically predictable sequence, defining six evolutionary stages, according to Braak et al.1,5
Genetic studies show that mutations in SNCA, PARKIN, UCHL-1, PINK1, DJ-1 and LRKK2 are the origin of familial cases of PD, although they account only for 5–10% of patients. The majority of PD cases are idiopathic.3,6-8 Even if familial and idiopathic forms of the disease differ on several clinical aspects, it is clear that common molecular pathways underlie neurodegeneration. These include oxidative stress, mitochondrial dysfunction, energy production imbalance and disruption of the ubiquitin-proteasome system. Gene expression profiling studies in different brain areas from sporadic PD patients have widely shown transcriptome deregulation affecting these pathways.9-16
MicroRNAs (miRNAs) and other small non-coding RNAs (sncRNAs) are post-transcriptional gene expression regulators, playing key roles in neuronal development, plasticity and disease. A strong body of evidence suggests that miRNA dysregulation underlies neurodegenerative processes. In PD, abnormal function of three miRNAs has been shown. For instance, the expression of miR-133b, a miRNA essential in the differentiation and maintenance of dopaminergic neurons in the SN, was reduced in midbrain samples from patients with PD.17 In addition, disruption of the binding site for miR-433 in the FGF20 gene has been associated to PD, and FGF20 overexpression has been shown to correlate with higher SNCA expression levels.18 Besides, miR-7, which modulates SNCA, was shown to be downregulated in a PD mouse model.19 Our own recent data indicate that downregulation of miR-34b/c occurs in several brain areas of PD patients and further suggest that miR-34b/c downregulation underlie early mitochondrial dysfunction in PD.20 Here we have further profiled sncRNAs in PD brains and found that upregulation of a small vault RNA (that we have named svtRNA2-1a) is a widespread event in PD brains. We have characterized svtRNA2-1a biogenesis, and analyzed the consequences of its deregulation in the neuronal transcriptome and physiology. Our results suggest that svtRNA2-1a deregulation could contribute to initial PD pathogenesis.
Results
svtRNA2-a is upregulated at advanced stages of PD
In a first attempt to identify small non-coding RNAs (sncRNAs) with a possible role in PD, we focused on amygdala (AM) from PD patients at motor stages of the disease (stages 4 and 5). At those stages, most individuals have crossed the threshold to the symptomatic phase of the illness, which is characterized by robust affectation of the substantia nigra (SN) and the AM. We profiled miRNA expression in the amygdala of 11 PD patients using a commercial miRCURY™ array (Exiqon) that contained all miRNAs present in the miRBase miRNA registry release 8.1 and a series of un-annotated sequences corresponding to either experimentally reported or predicted sncRNAs. Each individual case RNA sample was hybridized against a pool made of equivalent quantities of RNA from six control individuals (Table S1). Based on this array, we were able to identify two miRNAs (miR-34b and miR-34c-5p) that were significantly downregulated in PD patients.20 Besides, there was a sncRNA upregulated in all PD amygdalas (miR-Plus 17885; Fig. S1), which was not annotated in miRBase release 8.1. MiR-Plus 17885 is encoded in chromosome 5q31.1, embedded in the gene coding for vault-RNA2-1 (vtRNA2-1). In the miRBase releases 10–16, this sRNA was annotated as hsa-miR-886-5p. Later miRbase releases considered this sRNA as a fragment of the vault RNA2-1 and it was no longer annotated as hsa-miR-886. Therefore we have named it small vault RNA2-1a (svtRNA2-1a), following the nomenclature used for the recently described svtRNAs derived from vtRNA1-1 (svtRNAa and svtRNAb).21 The upregulation of svtRNA2-1a was confirmed by RT-PCR analysis on the same PD patients and control individuals using Taqman®-specific assays and either U6B or RNU58B as reference sncRNA (Fig. S1).
We assessed the expression levels of svtRNA2-1a by rRT-PCR in a wider number of AM samples from symptomatic PD patients (Braak staging > 3) and control individuals (Table S1) and results confirmed a significant upregulation of svtRNA2-1a of approximately 1.5-fold in AM from PD compared with control AM (Fig. 1A; Fig. S2). To gain insights into the mechanism leading to svtRNA2-1a upregulation, we determined the expression of its precursor vtRNA2-1. We could not detect significant changes in vtRNA2-1 expression in AM samples of patients with PD at motor stages, compared with control individuals, using custom TaqMan assays (Fig. S3). Thus, svtRNA2-1a overexpression might be linked with regulation of its biogenesis from vtRNA2-1 rather than an increase in the expression of vtRNA2-1.
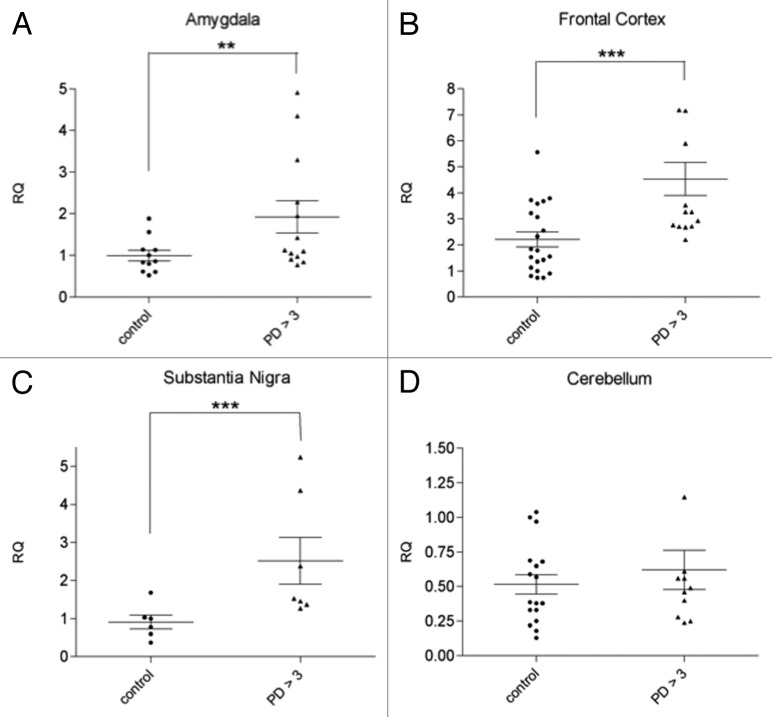
Figure 1. svtRNA2-1a expression in different brain regions from control individuals and PD cases stages 4 and 5 (motor stages of PD). svtRNA2-1a levels are shown in the amygdala (A), substantia nigra (B), frontal cortex (C) and cerebellum (D). svtRNA2_1 expression levels are referred to a control sample, for relative quantification (RQ). Plots show individual and mean RQ ± SEM (*, p < 0.05; **, p < 0.01; *** using a linear mixed effects model, LMM).
We next evaluated if svtRNA2-1a upregulation in PD symptomatic cases was extensive to other areas of the brain with different degrees of neuropathological affectation. rRT-PCR analysis revealed a significant upregulation of svtRNA2-1a of approximately 2.5-fold in the SN of symptomatic PD patients (Fig. 1B; Table S1, Fig. S2). In the frontal cortex (FC), an area that presents mild neuropathological alterations, we also observed an increase on svtRNA2-1a levels of approximately 2-fold in PD cases compared with control individuals (Fig. 1C; Table S1, Fig. S2). Finally, we could not detect significant changes in the expression levels of svtRNA2-1a in the cerebellum (CB) of PD patients, a structure with scarce lesions in PD (Fig. 1D; Table S1, Fig. S2). These results suggest that the extent of upregulation of svtRNA2-1a does not strictly correlate with the severity of the neuropathological lesions.
svtRNA2-a upregulation is an early event in PD that is not related to drug therapies
The severity of the intraneuronal lesions increases steadily along the course of the disease. The alterations develop, to some degree, even in the brain stem of persons whose clinical protocols refer no disease-associated motor dysfunctions.22,23 These incidental PD cases represent pre-symptomatic PD and are classified as Braak stages < 3. In order to further assess if svtRNA2-1a upregulation correlates with disease progression, we analyzed the expression of svtRNA2-1a in pre-symptomatic PD cases (Braak stages 1–3; Table S1, Fig. S2). At pre-clinical cases the AM starts to present subtle neuropathological lesions, whereas the FC is not affected. Using Taqman rRT-PCR assays we were able to detect a significant increase in the expression of svtRNA2-1a of approximately 2-fold in the AM (Fig. 2A) of pre-motor PD cases compared with control individuals but no significant changes were detected in the FC of pre-clinical cases (Fig. 2B). These results suggest that svtRNA2-1a upregulation is an early event in PD, at least in the AM. Importantly, none of these pre-symptomatic patients had received any PD-related treatment since they had not suffered from motor symptoms, and neuropathological changes were found incidentally in the course of the pathological examination. Therefore, the possibility of PD related-drug side effects on gene expression can be excluded.
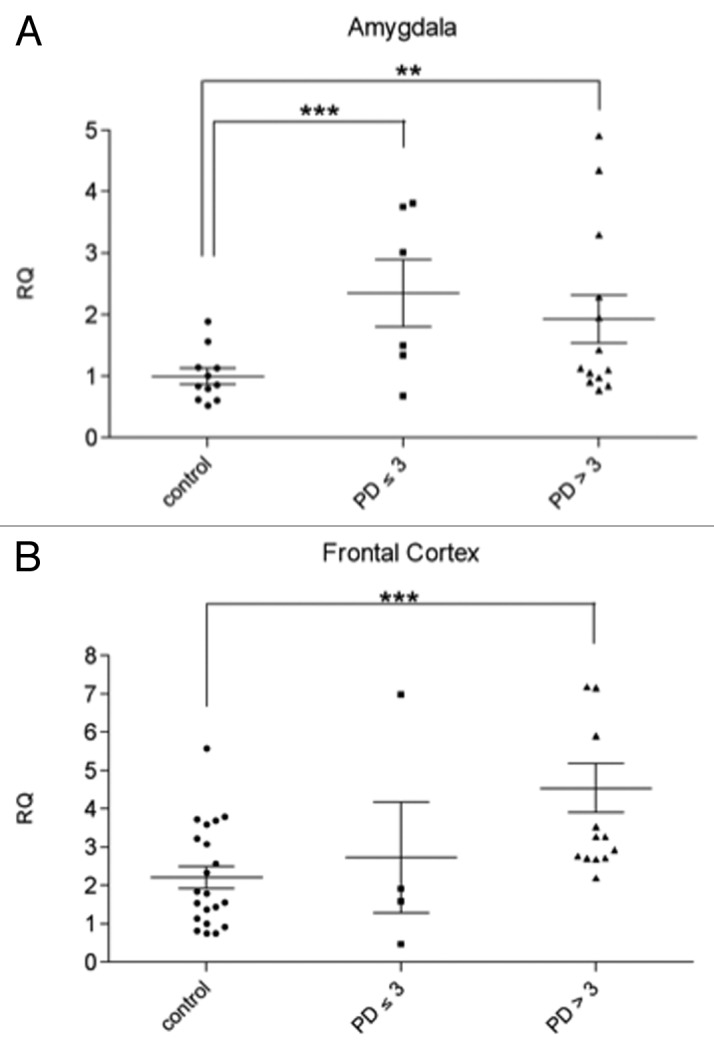
Figure 2. svtRNA2-1a expression in brain samples of pre-motor PD cases (stages 1–3) svtRNA2-1a expression levels in control, pre-motor PD (stages 1, 2 and 3) and clinical PD (stages 4 and 5) in the amygdala (A) and frontal cortex (B). Expression levels are referred to a control sample, for relative quantification (RQ). Plots show individual and mean RQ ± SEM (*, p < 0.05; **, p < 0.01 using a linear mixed effects model).
svtRNA2-a is enriched in Ago2 complexes and its biogenesis is dependent on Dicer
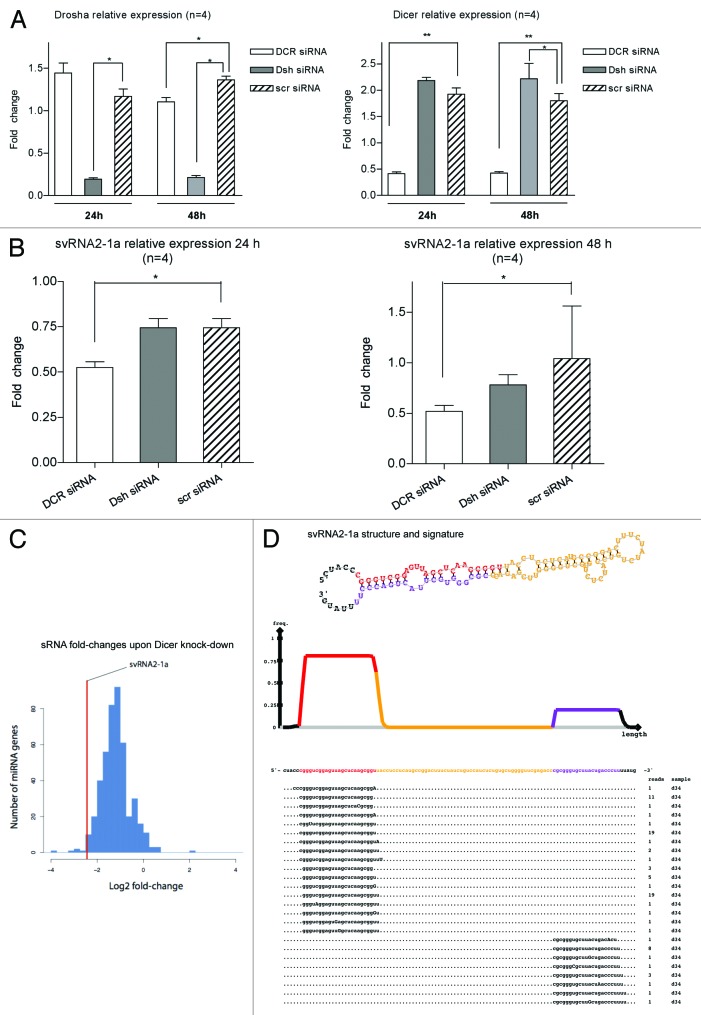
Recently, vtRNA1-1 was shown to produce svtRNAs in a Dicer-dependent and Drosha-independent process.21 Thus, we assessed whether svtRNA2-1a biogenesis occurred through an analogous mechanism. HeLa cells were depleted from Dicer or Drosha, using specific siRNAs, or a scrambled siRNA in four independent experiments; lipofectamine-treated cells were used as an additional negative control. RNA was extracted 24 and 48 h post-transfection, and both Dicer and Drosha expression were assessed by rRT-PCR to evaluate the extent of the knockdown. Dicer and Drosha expression levels were significantly reduced by 80%, compared with cells transfected with a scrambled control siRNA, 24 and 48 h after transfection (Fig. 3A). At the same time points, the expression of svtRNA2-1a was significantly decreased upon Dicer depletion, but not upon Drosha depletion (Fig. 3B). These results indicate that svtRNA2-1a biogenesis is dependent on Dicer but not on Drosha, confirming a biogenesis process similar to that of svtRNAa.21 Furthermore, we analyzed publicly available high-throughput small RNA sequencing data in control MCF7 cells and MCF7 cells depleted of Dicer.24 The normalized number of reads annotated as svtRNA2-1a, was significantly smaller (72%) in Dicer depleted cells compared with control cells supporting the involvement of Dicer in svtRNA2-1a generation. From the 450 miRNAs that could be robustly quantified in this study, svtRNA2-1a is among the top 10 sncRNAs whose expression is most affected by Dicer knockdown (Fig. 3C). The precise alignment of sequencing reads corresponding to vtRNA2-1 shows a strongly biased pattern toward two regions—the mature sncRNA of one arm of the stem (svtRNA2-1a) and its counterpart from the minor strand (svtRNA2-1a*), which is consistent with a Dicer-dependent biogenesis (Fig. 3D). A similar pattern was detected when analyzing 94 carefully quality filtered publicly available human sRNA high-throughput sequencing data sets [Gene Expression Omnibus (GEO) database]. Of the ~460,000 sequenced RNAs which we could trace to svtRNA2-1a with high confidence, ~420,000 (92%) correspond exactly to major strand, and ~35,000 (8%) correspond exactly to the minor strand (Fig. S4). The position of the strands have the typical 2 nt 3′ overhang, which is indicative of Dicer processing. Overall, we detected the major strand in 87 out of 94 data sets, while the minor strand was detected in 79 of them. These data suggest that vtRNA2-1 is a natural substrate of Dicer in different human cells and tissues.
Figure 3. svtRNA2-1a biogenesis is dependent on Dicer. (A) Plots show relative expression of Dicer and Drosha assessed by rRT-PCR. Mean fold change ± SEM, 24 and 48 h after transfection of specific siRNAs against Dicer (DCR), Drosha (Dsh) or a scrambled siRNA (scr); all values are referred to lipofectamine-only treated (n = 4; *, p value < 0.05, using LMM). (B) Plots show relative expression of svtRNA2-1a assessed by rRT-PCR. Mean fold change ± SEM 24 h after transfection of specific siRNAs against Dicer (DCR), Drosha (Dsh) or a scrambled siRNA (scr), lipofectamine only treated cells were used as the reference (n = 4; *, p value < 0.05 using LMM). Graphs show mean fold change (n = 4) 24 h after either Dicer or Drosha depletion. (C) Fold-changes in miRBase miRNAs after Dicer silencing in MCF-7 cell line, measured by high-throughput sequencing. The line indicates the reduction fold change for svtRNA2-1a. (D) VTRNA2-1 structure and putative biogenesis of svtRNA2-1a by Dicer cropping. SvtRNA2-1 sequencing reads alignment to vtRNA2-1 precursor, in a 34 d old small RNA sequencing data set, of the human brain. VTRNA2-1 secondary structure is shown and the different regions that define a typical miRNA precursor are highlighted in colors. The major strand (svtRNA2-1a) is shown in red, the minor strand in purple (svtRNA2-1a*) and the intervening loop in yellow. The density plot shows the distribution of sequencing reads. Below, the positions of individual sequences are shown. The column labeled “reads” indicate how many times the sequence was detected.
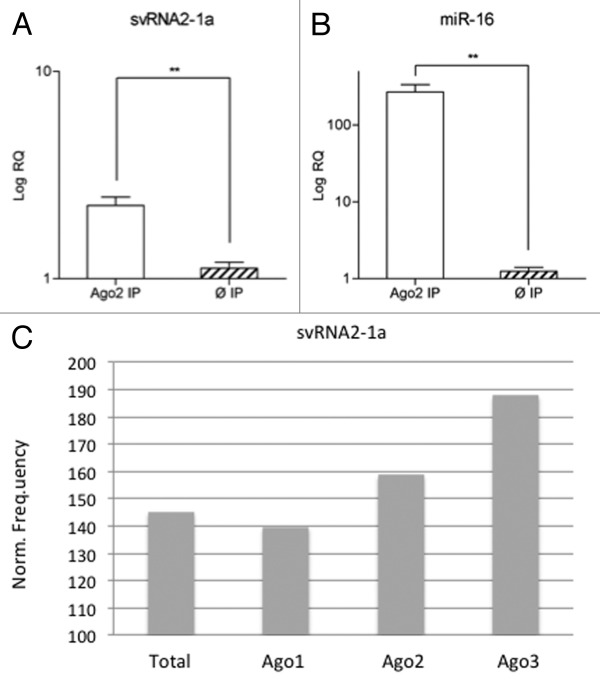
To assess a possible role of svtRNA2-1a as a silencing sncRNA in differentiated neuronal cells, we evaluated whether svtRNA2-1a was present in endogenous Ago2 complexes. For this, endogenous Ago2 was immunoprecipitated (IP) in SH-SY5Y neuronal cells. Differentiation to a post-mitotic dopaminergic phenotype was performed using retinoic acid (RA) and phorbol-12-myristate 13-acetate (TPA) as described in reference 20. Anti-RMC antibody raised against the C4 protein of the complement system in rat/mouse was used for control IP. Total RNA was isolated from the Ago2 and control IPs in three independent experiments and the expression of svtRNA2-1a and miR-16 (used as a positive control) was assessed by rRT-PCR Taqman assays. We detected a significant enrichment of svtRNA2-1a and miR-16 levels in Ago2 IPs (Fig. 4A and B). In addition, we evaluated the presence of svtRNA2-1a in different types of Ago immunocomplexes in human THP-1 monocytic cells, using publicly available small-RNA sequencing data sets.25 SvtRNA2-1a was found in Ago1, Ago2 and Ago3 immunocomplexes, being specially enriched in Ago3 (Fig. 4C). These results confirm the presence of svtRNA2-1a in Ago complexes and, therefore, suggest the plausible role of this sncRNA as a modulator of gene expression.
Figure 4. svtRNA2-1a is enriched in Ago complexes. (A and B) Expression of svtRNA2-1a in Ago2 immunocomplexes in SH-SY5Y cells. Plot shows mean Log RQ (relative quantity) ± SEM, for svtRNA2-1a (A) and miR-16 (B) assessed by rRT-PCR in Ago2-immunoprecipitates or control-immunoprecipitates in three independent immunoprecipitation experiments (**, p < 0.01). (C) svtRNA2-1a distribution in different Ago proteins. The expression of svtRNA2-1a was determined in immunoprecipitated Ago1, Ago2 and Ago3 complexes and total cell extracts from monocytic THP-1 human cells, using public sncRNA sequencing data. Norm frequency indicates the normalized frequency calculated as freq. svtRNA2-1a/freq. miRNAs * 10E6.
SvtRNA2-1a mimic overexpression induces subtle transcriptomic changes in SH-SY5Y cells
To gain insight into the pathways that could be altered by svtRNA2-1a deregulation, as well as to identify putative target transcripts, we assessed gene expression changes upon svtRNA2-1a overexpression in differentiated SH-SY5Y cells, using Agilent Sure print G3 human GE 8 × 60 k microarrays, which include not only updated transcriptome databases for mRNA targets but also probes for long intergenic non-coding RNAs (lincRNAs).
Differentiated SH-SY5Y cells were transiently transfected with either a specific svtRNA2-1a mimic or a mimic scrambled sequence (scr) in five independent experiments. Lipofectamine only (Ø) treated cells were used as an additional control. RNA was isolated 48 h after transfection and upregulation of svtRNA2-1a was confirmed by rRT-PCR (Fig. S5).
After normalization, data analysis and statistical filtering (expression fold change ≥ 1.2 or ≤ 0.83 when compared with both “scrambled mimic” and “lipofectamine only” controls; and adjusted p value ≤ 0.25; plus no expression differences between “scrambled” and “lipofectamine only” controls) we observed a set of 103 moderately deregulated transcripts. Out of these 103 transcripts, 41 corresponded to lincRNAs (Tables 1 and 2). Most lincRNA were upregulated, and considering the protein coding genes, 55 were upregulated and six downregulated. These data suggest either that the mild gene upregulation detected at the time-point post-transfection examined may be secondary to svtRNA2-1a silencing activity or that the regulation of gene expression mediated by svtRNA2-1a does not necessarily involve target mRNA decay. Considering the six downregulated genes, only SOX4 was identified as a putative svtRNA2-1a target by MirWalk26 and PITA27 prediction algorithms. Among the downregulated genes, SPARCL1 and TXNIP expression are decreased in several studies according to two publicly available databases for gene expression profiling in PD (ncascr.griffith.edu.au/pdreview/2008/ and bioportal.kobic.re.kr/PDbase/, refs. 28 and 29). We validated a slight TXNIP downregulation by rRT-PCR, while we failed to amplify SOX4 (Table 3). Overall, the present data suggest that gene expression deregulation by svtRNA2-1a is subtle at the RNA level.
Table 1. Deregulated protein-coding transcripts upon svRNA2-1a overexpression (in bold selected for rRT-PCR validation).
| Gene Name | F.C. svRNA2-1a vs (SCR +Ø) | adj.P.Val. svRNA2-1a vs (SCR +Ø) | F.C. SCR vs Ø | adj.P.Val. SCR vs Ø |
|---|---|---|---|---|
| A_33_P3237704 |
1.2330 |
0.2388 |
-1.0524 |
0.9999 |
| A_33_P3265941 |
1.2413 |
0.2475 |
-1.0052 |
0.9999 |
| A_33_P3365963 |
1.2523 |
0.2498 |
-1.0704 |
0.9999 |
| A_33_P3371144 |
1.2266 |
0.2475 |
-1.0810 |
0.9999 |
| A_33_P3391536 |
1.4585 |
0.2475 |
-1.0405 |
0.9999 |
| A_33_P3414017 |
1.2375 |
0.2475 |
-1.0293 |
0.9999 |
| APOL1 |
1.4084 |
0.2475 |
-1.1972 |
0.9999 |
| ATN1 |
1.2024 |
0.2475 |
-1.0583 |
0.9999 |
|
ATP1A3 |
1.2498 |
0.1248 |
-1.0403 |
0.9999 |
| C20orf141 |
1.2125 |
0.2475 |
-1.0716 |
0.9999 |
| C9orf173 |
1.2646 |
0.2475 |
-1.0650 |
0.9999 |
| CA6 |
1.4151 |
0.2475 |
-1.2019 |
0.9999 |
| CASKIN1 |
1.2015 |
0.2475 |
-1.0372 |
0.9999 |
| CDC14C |
1.2300 |
0.2475 |
-1.0771 |
0.9999 |
| CENPP |
1.2177 |
0.2201 |
1.0647 |
0.9999 |
| CHST6 |
1.2243 |
0.2475 |
-1.0706 |
0.9999 |
|
CRTC1 |
1.3100 |
0.1782 |
-1.1205 |
0.9999 |
| DKFZP547L112 |
1.3196 |
0.2475 |
-1.0859 |
0.9999 |
| ENST00000324659 |
1.2848 |
0.2475 |
-1.0479 |
0.9999 |
| ENST00000378223 |
1.2925 |
0.2475 |
1.0058 |
0.9999 |
| ENST00000390293 |
1.2509 |
0.2475 |
-1.0863 |
0.9999 |
| FLJ43860 |
1.2789 |
0.2475 |
-1.0507 |
0.9999 |
| GPSM1 |
1.2072 |
0.2475 |
-1.0222 |
0.9999 |
| GYPA |
1.2439 |
0.2475 |
1.0353 |
0.9999 |
| HBZ |
1.2560 |
0.2475 |
-1.0780 |
0.9999 |
| HIST1H2AM |
1.2034 |
0.2475 |
1.0679 |
0.9999 |
| HNRNPUL1 |
1.2248 |
0.2475 |
-1.0142 |
0.9999 |
| HOXA9 |
1.2020 |
0.2475 |
1.0048 |
0.9999 |
| IQSEC2 |
1.2369 |
0.2475 |
-1.0758 |
0.9999 |
| KCP |
1.2280 |
0.2475 |
-1.0651 |
0.9999 |
| KIRREL2 |
1.2357 |
0.2475 |
-1.0628 |
0.9999 |
|
LHX3 |
1.3588 |
0.2475 |
-1.0654 |
0.9999 |
| LOC100129196 |
1.2054 |
0.2201 |
-1.0215 |
0.9999 |
| LOC100132966 |
1.2777 |
0.2475 |
-1.0784 |
0.9999 |
| LOC388152 |
1.2027 |
0.2475 |
-1.0788 |
0.9999 |
| LY9 |
1.2376 |
0.2475 |
1.0250 |
0.9999 |
|
LYNX1 |
1.2769 |
0.2475 |
-1.0365 |
0.9999 |
| NKX1–2 |
1.2990 |
0.2475 |
-1.1392 |
0.9999 |
| NOBOX |
1.2333 |
0.2388 |
1.0089 |
0.9999 |
| OGFR |
1.2302 |
0.2475 |
-1.0343 |
0.9999 |
| OR5L2 |
1.2448 |
0.2475 |
-1.0622 |
0.9999 |
| OSCAR |
1.2536 |
0.2475 |
-1.0760 |
0.9999 |
| PLK1 |
1.2115 |
0.2475 |
1.0039 |
0.9999 |
| PRKAG3 |
1.2568 |
0.2419 |
-1.0628 |
0.9999 |
| SCGB3A1 |
1.2214 |
0.2475 |
-1.0600 |
0.9999 |
| SF3A2 |
1.2295 |
0.2475 |
-1.0376 |
0.9999 |
| SIGLEC15 |
1.3126 |
0.2475 |
1.0176 |
0.9999 |
| SLC34A3 |
1.2093 |
0.2475 |
-1.0674 |
0.9999 |
|
SNCB |
1.2263 |
0.2475 |
-1.0533 |
0.9999 |
| SP8 |
1.3189 |
0.2475 |
-1.1346 |
0.9999 |
| THBS1 |
1.2325 |
0.2475 |
-1.0773 |
0.9999 |
| TRIM3 |
1.2424 |
0.2475 |
-1.0716 |
0.9999 |
| TUBA3D |
1.2361 |
0.2475 |
1.0011 |
0.9999 |
| TUSC1 |
1.2405 |
0.2475 |
-1.0678 |
0.9999 |
| ZNF843 |
1.3978 |
0.2388 |
-1.0643 |
0.9999 |
| LCA5 |
-1.2092 |
0.2475 |
-1.0367 |
0.9999 |
| PURG |
-1.2425 |
0.2475 |
-1.1683 |
0.9269 |
|
SOX4 |
-1.2189 |
0.2475 |
-1.0349 |
0.9999 |
|
SPARCL1 |
-1.2608 |
0.2475 |
-1.0658 |
0.9999 |
| TRDMT1 |
-1.2119 |
0.2475 |
1.0271 |
0.9999 |
| TXNIP | -1.2608 | 0.2475 | -1.0013 | 0.9999 |
In bold, predicted targets for svRNA2-1a.
Table 2. Deregulated linc-RNAs upon svRNA2-1a overexpression.
| Gene Name | F.C. svRNA2-1a vs (SCR +Ø) | adj.P.Val. svRNA2-1a vs (SCR +Ø) | F.C. SCR vs Ø | adj.P.Val. SCR vs Ø |
|---|---|---|---|---|
| lincRNA:chr20:25213650-25221750_F |
1.4051 |
0.2475 |
-1.2437 |
0.9909 |
| lincRNA:chr13:44760050-44767325_F |
1.3964 |
0.2415 |
-1.0620 |
0.9999 |
| lincRNA:chr16:11481046-11484311_F |
1.3409 |
0.2475 |
1.0205 |
0.9999 |
| lincRNA:chr14:56163972-56175072_R |
1.3072 |
0.2475 |
-1.0646 |
0.9999 |
| lincRNA:chr3:36985666-36986208_F |
1.2990 |
0.2475 |
-1.0208 |
0.9999 |
| lincRNA:chr5:133837051-133848098_R |
1.2950 |
0.2475 |
-1.0874 |
0.9999 |
| lincRNA:chr19:17187375-17194450_R |
1.2946 |
0.2475 |
-1.0792 |
0.9999 |
| lincRNA:chr10:6779694-6889494_R |
1.2917 |
0.2475 |
-1.0267 |
0.9999 |
| lincRNA:chr3:185544131-185550706_F |
1.2844 |
0.2475 |
-1.0207 |
0.9999 |
| lincRNA:chr2:218461580-218481005_R |
1.2831 |
0.2388 |
-1.0517 |
0.9999 |
| lincRNA:chr2:238532786-238566636_R |
1.2786 |
0.2475 |
-1.0347 |
0.9999 |
| lincRNA:chr18:33160711-33161067_F |
1.2750 |
0.2475 |
-1.0659 |
0.9999 |
| lincRNA:chr8:145986196-145991421_F |
1.2728 |
0.2475 |
-1.0227 |
0.9999 |
| lincRNA:chr5:167692897-167704922_F |
1.2708 |
0.2475 |
-1.0447 |
0.9999 |
| lincRNA:chr22:46476224-46493888_F |
1.2660 |
0.2475 |
-1.0569 |
0.9999 |
| lincRNA:chr11:70606952-70667252_F |
1.2656 |
0.2475 |
-1.0534 |
0.9999 |
| lincRNA:chr2:241928152-241929111_R |
1.2651 |
0.2475 |
-1.0430 |
0.9999 |
| lincRNA:chr6:72033379-72059179_F |
1.2647 |
0.2475 |
-1.0591 |
0.9999 |
| lincRNA:chr12:114196867-114203667_F |
1.2645 |
0.2201 |
-1.0072 |
0.9999 |
| lincRNA:chr6:32862169-32868745_F |
1.2633 |
0.2475 |
-1.1159 |
0.9999 |
| lincRNA:chrX:16499154-16509904_F |
1.2603 |
0.2475 |
-1.0621 |
0.9999 |
| lincRNA:chr2:193943030-194203155_R |
1.2558 |
0.1817 |
-1.0219 |
0.9999 |
| lincRNA:chr4:4545549-4583049_F |
1.2542 |
0.2475 |
-1.0912 |
0.9999 |
| lincRNA:chrX:45598006-45710454_F |
1.2306 |
0.1817 |
-1.0474 |
0.9999 |
| lincRNA:chr5:52531518-52727218_R |
1.2289 |
0.2475 |
1.0002 |
0.9999 |
| lincRNA:chr4:7132149-7143349_F |
1.2277 |
0.2475 |
-1.0195 |
0.9999 |
| lincRNA:chr9:110102958-110105481_F |
1.2273 |
0.2475 |
-1.0048 |
0.9999 |
| lincRNA:chr2:64412771-64525721_R |
1.2255 |
0.2475 |
-1.0483 |
0.9999 |
| lincRNA:chr4:84135426-84176451_F |
1.2245 |
0.2475 |
-1.0344 |
0.9999 |
| lincRNA:chr12:2035164-2049639_R |
1.2236 |
0.2475 |
1.0187 |
0.9999 |
| lincRNA:chr11:508000-516800_F |
1.2234 |
0.2201 |
-1.0984 |
0.9999 |
| lincRNA:chr1:41956288-41971088_R |
1.2160 |
0.2475 |
-1.0549 |
0.9999 |
| lincRNA:chr12:76015433-76261508_F |
1.2151 |
0.1817 |
-1.0271 |
0.9999 |
| lincRNA:chr12:9480083-9506383_R |
1.2145 |
0.2475 |
-1.0082 |
0.9999 |
| lincRNA:chr7:106224664-106257564_F |
1.2119 |
0.2475 |
1.0095 |
0.9999 |
| lincRNA:chr18:33466152-33528357_F |
1.2089 |
0.2475 |
-1.0835 |
0.9999 |
| lincRNA:chr10:54147394-54152519_F |
1.2086 |
0.2431 |
-1.0430 |
0.9999 |
| lincRNA:chr5:54162493-54208143_R |
1.2082 |
0.2475 |
-1.0220 |
0.9999 |
| lincRNA:chr9:16042794-16061320_F |
1.2039 |
0.2475 |
-1.0455 |
0.9999 |
| lincRNA:chr6:36084247-36091829_F |
1.2026 |
0.2475 |
-1.0293 |
0.9999 |
| lincRNA:chr6:43819493-43820070_R | 1.2009 | 0.2388 | -1.0077 | 0.9999 |
Table 3. rRT-PCR validation of selected protein-coding transcripts deregulation upon svRNA2-1a overexpression.
| svRNA2-1a vs [SCR+Ø] | SCR vs Ø | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| |
Housekeeping genes |
|
Housekeeping genes |
||||||
| |
MRIP |
POLR2J |
|
MRIP |
POLR2J |
||||
|
GENE |
FC |
p value |
FC |
p value |
GENE |
FC |
p value |
FC |
p value |
| SPARCL1 |
0.989 |
0.842 |
0.977 |
0.670 |
SPARCL1 |
0.951 |
0.318 |
0.973 |
0.689 |
|
TXNIP |
0.908 |
0.047 |
0.897 |
0.091 |
TXNIP |
0.929 |
0.195 |
0.951 |
0.549 |
| ATP1A3 |
0.847 |
0.135 |
0.803 |
0.163 |
ATP1A3 |
0.963 |
0.764 |
1.117 |
0.549 |
| CRTC1 |
0.923 |
0.036 |
0.964 |
0.689 |
CRTC1 |
0.971 |
0.484 |
0.994 |
0.999 |
| LYNX1 |
0.955 |
0.318 |
0.998 |
0.999 |
LYNX1 |
0.954 |
0.369 |
0.977 |
0.842 |
| SNCB | 1.145 | 0.029 | 1.116 | 0.318 | SNCB | 1.076 | 0.272 | 1.142 | 0.272 |
SvtRNA2-1a levels may modulate neuronal differentiation and maintenance
In order to gain insight into the pathways that may be regulated by svtRNA2-1a, we evaluated function enrichment using the Ingenuity Pathway Analysis (IPA) tool for both the significantly deregulated genes upon svtRNA2-1a overexpression in the array, and separately, the predicted targets for svtRNA2-1a (Table S2). To predict svtRNA2-1, a putative targets we used the prediction tool MirWalk (www.ma.uni-heidelberg.de/apps/zmf/mirwalk/index.html),26 considering all the genes with predicted target sites in the 3′UTR, by at least two different algorithms from the following: miRWalk, miRDB, miRanda, PITA, Targetscan and MirBase. In both cases, the most significant biological functions involved are related to the maintenance and differentiation of neuronal cells and the nervous system (Tables S3 and S4).
To evaluate if the amount of svtRNA2-1a was linked to developmental processes of the human brain we analyzed the expression pattern of svtRNA2-1a in high-throughput sequencing data sets of human frontal cortex during post-natal development and aging.30 We found that both the major and minor strands reach their highest expression in the first year after birth, with progressively lower expression afterwards (Fig. 5B), reinforcing the idea that svtRNA2-1a levels may participate in neuronal differentiation and maintenance.
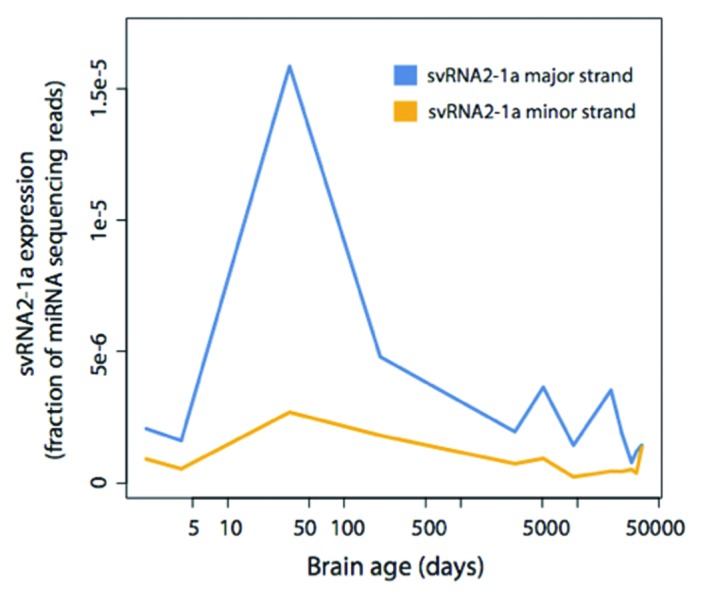
Figure 5. svtRNA2-1a expression in human brain development and aging. svtRNA2-1a expression in the human brain detected by sncRNA sequencing. The time points are: 2 d after birth, 4 d, 34 d, 204 d, 8 y, 13 y, 25 y, 53 d, 66 y, 88 y and 98 y. Expression has been normalized to the number of miRNA sequencing reads.
To assess a possible role of svtRNA2-1a deregulation in neuronal maintenance, we determined basal viability in cells transiently transfected with a sv-RNA1-2a mimic, using the [3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] (MTT) assay. We found decreased MTT metabolization in differentiated SH-SY5Y cells overexpressing a svtRNA2-1a mimic vs. those overexpressing a scrambled-control sequence, at different time-points post-transfection (Fig. 6), suggesting an impairment in the activity of reductase enzymes. Decreased MTT metabolization has been typically linked to mitochondrial impairment and oxidative stress, and cell death. However, we did not detect obvious disruption of mitochondrial membrane potential, nor ROS generation after transfection with svtRNA2-1a mimics, using specific fluorescent dyes (data not shown). In addition, linked to svtRNA2-1a overexpression no obvious changes in cell density and morphology were detected, nor nuclear condensation was observed (data not shown). Thus, reduction of MTT metabolization in cells overexpressing svtRNA2-1a may reflect metabolic impairment rather than cell death linked to mitochondrial dysfunction. In fact, it has been shown that although mitochondrial succinate dehydrogenase can reduce MTT, microsomal enzymes are the main contributors to total cellular MTT reduction. Furthermore, cellular MTT reduction is affected by glucose availability,31,32 thus supporting the view that decreases in MTT reduction driven by svtRNA1-2a overexpression could reflect glycolytic metabolism impairment.
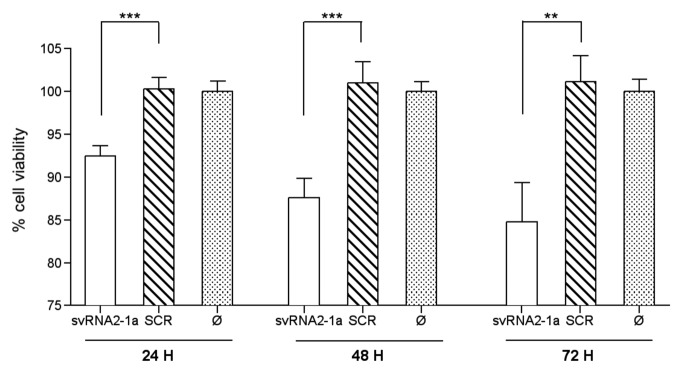
Figure 6. svtRNA2-1a overexpression reduces neuronal basal viability. Plot shows relative cell viability assessed with MTT assays at 24, 48 and 72 h after transfection of either svtRNA2-1a mimic or a scrambled miRNA mimic (SCR), using lipofectamine only (Ø) treated cells as negative control and reference. Data represent mean basal viability ± SEM [n = 6; (**, p < 0.01, ***, p < 0.001 using t-test)].
Discussion
Previous studies suggest that miRNA deregulation contributes to neurodegenerative processes.17,18,33-37 Different studies, including our own recent data, suggest that altered miRNA expression profiles underlie PD pathogenesis.17,19,20 Here we have identified a vault RNA-derived small RNA, svtRNA2-1a, which is consistently upregulated in several PD brain regions, differently affected at the neuropathological level. Our data suggest that svtRNA2-1a could be synthesized as a non-canonical miRNA, and modulate gene-expression networks involved in neuronal maintenance.
SvtRNA2-1a was originally registered as miR-886-5p, in the miRBase release 10, because it was captured, together with miR-886-3p in high-throughput sequencing and the two mature miRNAs formed a stem in a predicted stem-loop hairpin structure, which is a signature for a miRNA precursor.38,39 SvtRNA2-1a is embedded in the gene coding for the vault RNA VTRNA2. The human vtRNAs are 88 or 98 nt in length, are RNA polymerase III (Pol III) transcripts and have a stem-loop secondary structure. All these features are shared by pre-miR-886, and for this reason it was recently proposed to be renamed as vtRNA2-1.40,41 VtRNAs are components of the vault complex, a hollow, barrel-shaped ribonucleoprotein complex that is thought to play a role in cellular resistance to cancer therapeutic (for a review see ref. 42). However, the exact role of vtRNAs, as well as the vault complex, is not yet clear. VtRNAs, are ubiquitously expressed, and they can all be detected free in the cytoplasm,43 thus suggesting they may all exert some function on their own. This is especially relevant in the case of vtRNA2-1, for which recent studies show a poor stable association with the vault complex.44
Our experimental data and evidences from sRNA high-throughput sequencing upon Dicer depletion in MCF7 cells,24 show that svtRNA2-1a biogenesis is dependent on Dicer, but not on Drosha, unlike canonical miRNAs. Furthermore, our analyses on the alignment of vtRNA2-1 sequencing reads indicate a pattern consistent with a Dicer-dependent biogenesis. Thus svtRNA2-1a generation is analogous to that described for svtRNAa and svtRNAb from vtRNA1.21 A non-canonical Drosha-independent biogenesis pathway has also been reported for mitrons, short introns, which after splicing and de-branching, form pre-miRNA-like molecules that are a source of pre-miRNA molecules.45-46 A recent study reported low efficiency for the processing of the stem-loop precursor to the mature svtRNA2-1a and svtRNA2-1a* (miR-886-5p and miR-886-3p) in epithelial cells.44 However, the mature minor strand (svtRNA2-1a*/hsa-miR-886-3p) is detected by northern blot in HS5 cells, a human stromal cell line.48 In addition, our results suggest that in PD, svtRNA2-1a processing from vtRNA2-1 is enhanced. These data suggest that the efficiency in svtRNA2-1a biogenesis may depend on the cellular context. In addition, more sensitive techniques such as rRT-PCR49-51 and sRNA high-throughput sequencing (GEO sncRNA sequencing data sets) show consistent detection of both of the mature forms across different cell lines, tissues and biological conditions, which is in agreement with a biological role of these sRNAs.
We observed a significant increase in the expression of svtRNA2-1a in the AM 1.5-fold change, FC 2.2-fold change, SN 2.6-fold change of symptomatic PD patients; while no changes in the svtRNA2-1a levels where detected in the CB. Similar deregulation levels for other silencing small RNAs have been detected in PD20 and Huntington disease,52 which were detrimental for neuronal viability. In clinical cases of PD, several abnormalities have been detected in areas virtually depleted of Lewy inclusions (FC and CB), including mitochondrial dysfunction, oxidative stress and synaptic pathology.16,53-58 This suggests that svtRNA2-1a upregulation may be linked to complex molecular pathology rather than to the distribution of Lewy inclusions. Furthermore, our observations indicate that svtRNA2-1a upregulation occurs at early (pre-motor) stages of the disease, thus suggesting a possible role in the initial pathogenic events in PD.
It is worth mentioning that at these advanced stages of the disease, the SN is already depleted from approximately 60–70% of the dopaminergic neurons and, therefore, whether svtRNA2-1a upregulation occurs in the remaining neurons or in other cell types, such as glial cells, is an open question. Interestingly, predicted targets for svtRNA2-1a are significantly enriched in pathways involved in the differentiation and maintenance of neuroglia and, thus, svtRNA2-1a deregulation could contribute to glial cells dysfunction.
Recent studies indicate that infectious and inflammatory pathways underlie activation of vtRNA expression,40,59 therefore suggesting that svtRNA2-1a deregulation could be linked with expression alterations of its precursor. However, we did not observe significant upregulation of vtRNA2-1 in the same samples showing svtRNA2-1a overexpression. These results suggest that svtRNA2-1a deregulation is related with alterations in the maturation process from the precursor. The post-transcriptional regulation of miRNA expression levels has been reported for miR-138, which shows variable expression in different cell types without modifications of its precursor pre-miR-138-2.60 The strong Dicer dependence of svtRNA2-1a generation suggests that the amount and activity of this endonuclease could be a limiting factor for svtRNA2-1a biogenesis, both in physiological and pathological conditions. The Parkinson disease database (www2.cancer.ucl.ac.uk/Parkinson_Db2/index.php) indicates that Dicer is overexpressed in the FC and SN of patients with PD, which could contribute to svtRNA2-1a increases. However, it is worth mentioning that recent work has identified a battery of proteins that regulate Dicer processing activity60,61 and, thus, whether Dicer is particularly active in the brain areas showing svtRNA2-1a overexpression in PD, is an open question that deserves specific research.
Our results confirm that svtRNA2-1a is enriched in endogenous Ago complexes and, therefore, suggest a role for svtRNA2-1a in post-transcriptional gene regulation. Transcriptome profiling of differentiated SH-SY5Y cells overexpressing svtRNA2-1a showed subtle changes in gene expression, suggesting that mRNA degradation may not be central in svtRNA2-1a regulation of gene expression. In line with this, Bax is a target for svtRNA2-1a (miR-886-5p) that is repressed at the protein level, without affecting its mRNA expression.50 In addition, the analysis of small-RNA sequencing data in human monocytic cells suggests a preferential loading of svtRNA2-1a onto Ago3, which shows silencing activity at the translational level, without target mRNA slicing. Interestingly, it has been recently shown that pre-miR-886 (vtRNA2-1) has a tumor-suppressing role, through binding to PRKRA (protein kinase, interferon inducible double-stranded RNA-dependent activator).44 Thus, gene expression deregulation driven by svtRNA2-1a mimic overexpression in differentiated SH-SY5Y cells could reflect both downstream effects of svtRNA precursor (vtRNA2-1) and primary and secondary effects linked to svtRNA2-1a silencing.
The regulated expression of svtRNA2-1a in human brain development and aging are indicative of a role of this sncRNA in the central nervous system physiology. In the human superior frontal gyrus,30 the highest levels of svtRNA2-1a were detected at early postnatal developmental stages. After the first year, the amount of svtRNA2-1a decreased with low levels being detected at the oldest ages examined. These data suggest that high levels of svtRNA2-1a may modulate early developmental processes, while low levels may be required for neuronal maintenance and survival. Reinforcing this idea, pathway enrichment analysis among the svtRNA2-1a deregulated protein-coding genes and the predicted svtRNA2-1a targets identified nervous system development among the top biological functions. In addition, our observations indicate that svtRNA2-1a overexpression in differentiated SH-SY5Y impairs neuronal metabolism. It is worth noting that idiopathic PD is heralded by decreased glucose metabolism in different brain regions62-65 and, therefore, svtRNA2-1a overexpression could contribute to metabolic impairment in PD.
In conclusion, we have shown that svtRNA2-1a biogenesis is largely dependent on Dicer, in agreement with previous studies.21,44 Its enrichment in Ago immunocomplexes suggests a role for this svtRNA in post-transcriptional gene regulation. Our results further suggest that early deregulation of svtRNA2-1a expression in PD may contribute to the initial perturbations of gene expression networks, underlying cell functional impairment. A better understanding of the pathways regulated by svtRNA2-a, and also the mechanisms regulating its expression should provide new insights on the role of vtRNA in the central nervous system and also facilitate the identification of new targets for therapeutic approaches in PD.
Materials and Methods
Patient brain samples
Brain samples were obtained from the Institute of Neuropathology Brain Bank following the guidelines of the local ethics committee. The time between death and brain processing named post-mortem time or post-mortem delay, was between 3.00 and 11.00 h. Brains were removed from the skull and the left cerebral hemisphere, left cerebellum and alternate transversal sections of the brain stem were fixed in 4% buffered formalin. The rest of the brain and cerebellum was immediately cut into 1 cm-thick coronal sections. These and the alternate sections of the brain stem were frozen on dry ice and stored at -80°C until use. Neuropathological studies were performed on formalin-fixed paraffin embedded sections 4 micron-thick of 25 standard selected areas, which were processed for current neuropathological methods including hematoxylin and eosin, Klüver-Barrera and -amyloid, hyper-phosphorylated tau, -synuclein, ubiquitin, B-crystalin, TDP-43 and astroglial and microglial immunohistochemistry.
PD-related pathology was graded following Braak stagesCases with PD stages 4 and 5 (also named LBD stage 4 and 5) had suffered from parkinsonism and had been subjected to different treatments geared to control motor symptoms for variable period of time (cases 1–11, 18–19 and 21–23; Table S6); none of them had suffered from dementia. Cases with PD-related pathology stages 1–3 (LBD stages 1–3) had not suffered from motor symptoms, did not receive any specific therapy and the observation of LBs and LNs was an incidental feature at neuropathological examination (cases 12–17 and 20; Table S6). PD cases with additional pathology not related to synuclein, excepting those with a few neurofibrillary tangles in the entorhinal and transentorhinal cortices, were excluded from the study.
Age-matched control cases were chosen on the basis of the lack of neurological, metabolic and mental disorders, together with the lack of brain lesions, including the absence of neurofibrillary tangles, α-synuclein inclusions, TDP-43 abnormalities and lack of small vascular disease after neuropathological examination following the same protocol as the used for PD cases. A few diffuse β-amyloid plaques and a few neurofibrillary tangles in the entorhinal cortex were the only abnormality in some cases.
RNA isolation
Total RNA was isolated using miRNeasy (Quiagen) according to manufacturer’s instuctions. Briefly, to isolate RNA from cultured cells, cells were rinsed once with PBS 1x, then either lysed with 700 µL of Quiagen lysis buffer and protocol was followed up as specified or frozen at -80°C until isolation could be performed (never more than 1 mo). To isolate RNA from frozen brain samples, between 50–75 mg of tissue were homogenized using a Ultra Turrax T-8 homogenizer in 1 mL of Quiagen lysis buffer. Lysate was then frozen at -80°C and protocol was followed up within less than 2 wk.
RNA integrity and quality was assessed routinely with a Bioanalyser (Agilent), using the eukaryotic total RNA kit, according to manufacturer’s instructions. RNAs with RIN (RNA integrity number) below 5 were discarded.
Ago2 immunoprecipitation
Endogenous Ago2 IP, was performed as previously described,66 using an antibody against RMC as a negative control for the IP, in three independent experiments on differentiated SH-SY5Y neuronal cells. RNA was subsequently isolated from Ago2 immunocomplexes using phenol-chloroform. Briefly, 300 µL of phenol-chloroform were added to precipitated RNA, shacked and then centrifuged at maximum speed (17,000 g) for 15 min enough to separate the phases. The upper phase was then recovered (300 µL) and 700 µL of ethanol (100%) added, after shaking samples were left overnight at -20°C. RNA was recovered by centrifugation at full speed (17,000 g) and 4°C for 30 min. Remaining ethanol was eliminated by warming at 65°C for 20–30 s. RNA was then dissolved in RNase-free water and left in ice for approximately 1 h and then frozen at -80°C.
rRT-PCR
MiRNA-RT-PCR were performed using Taqman® miRNA-specific RT and rRT-PCR assays, following manufacturer’s instructions, starting with 20 ng of RNA per RT reaction, in an AB 7900HT Fast Real-Time PCR System. In the analysis of miRNA expression in brain areas, for each region all cases and controls were analyzed in the same rRT-PCR experiment. Each sample was run in quadruplicates when feasible, or in duplicates in two independent plates; and the real-time reaction repeated at least twice. All the specific taqman miRNA assays used are listed in Table S5.
For mRNA rRT-PCR, 2 µg of total RNA were treated with DNase (Ambion) according to manufacturer’s instructions. RNA was then quantified and 250 µg of DNase-treated RNA were retrotranscribed with a 1:1 mix of random hexamers and oligodT, using SuperScript III First-Strand Synthesis SuperMix for qRT_PCR (Invitrogen). cDNA was diluted 1:8 and 1 µL was used for RT-PCR reactions using Taqman®-specific gene expression assays, following manufacturer’s instructions in an AB 7900HT Fast Real-Time PCR System. Two independent RT reactions were performed for each sample, and then for each RT, samples were run in duplicates in two independent RT-PCR reactions. All the specific taqman mRNA assays used are listed in Table S5.
Relative quantification and statistical analysis of rRT-PCR data
Relative quantification (RQ) as shown in graphs was calculated with the 2-∆∆Ct method67 to compare expression values of a given miRNA or mRNA normalized to that of the housekeeping gene, two different reference genes were used: U6B and RNU58B for miRNA determinations, and POLR2J and M-RIP for mRNA quantifications. The RQ and their statistical significance were obtained from two different methods: a linear mixed effects model (LMM) as previously described20 that accounts for the different sources of variation derived from the experimental design and the Relative Expression Software Tool (REST ©).68
Expression arrays
MiRNA microarrays were performed and statistically analyzed by Exiqon using mercury LNA microarrays (based on Sanger database v.8.1). Each individual PD AM RNA sample was hybridized against a pool made of equitable quantities of all AM control samples; RNA quality was assessed with a Bioanalyser (Agilent).
Gene expression profiling was performed using one-color Agilent Sure print G3 human Gene Expression 8 × 60 k microarrays, which contain 60,000 probes, including 27,958 Entrez Gene RNAs and 7,419 lincRNAs. Briefly, 500 ng of total RNA from each sample were chemically labeled by dephosphorylating using calf intestinal alkaline phosphatase (CIP) and ligating Cyanine3-pCp by a T4-RNA ligase using Agilent mRNA Complete Labeling and Hyb Kit (p/n5190-0456). Labeled samples were dried and resuspended in 18 µl of nuclease-free water and co-hybridized with in situ hybridization buffer (Agilent) for 20 h at 55°C and washed at room temperature for 5 min in Gene Expression Wash Buffer 1 (Agilent) and 5 min at 37°C in Gene Expression Wash Buffer 2 (Agilent). The images were generated on a confocal microarray scanner (G2505B, Agilent) at 5 um resolution and quantified using Feature Extraction (Agilent). Extracted log2-transformed intensities were quantile normalized to make all data comparable.
Gene expression was compared between SH-SY5Y cells transiently transfected with a svtRNA2-1a mimic and control cells transfected with a scrambled mimic sequence (Table S6), or mock-transfected with lipofectamine. Successful transfection of svtRNA2-1a mimic was confirmed by rRT-PCR and RNA quality assessed with a Bioanalyser (Agilent). All RNA samples were individually hybridized. To assess differential expression, significance analysis of microarrays (SAM) was used.69 Results of SAM analysis were corrected for multiple testing according to the false discovery rate (FDR) method.70 Probes with FDR-adjusted p value below 25% and additionally a fold change exceeding 1.2 in absolute value were selected as the relevant ones. Final relative expression values were computed by taking the median log2 ratio of the respective probes per each mRNA. All statistical analyses were performed with Bioconductor project (version 2.3) in the R statistical environment.71
Computational analysis of deep sequenced svtRNA2-1a
Small RNAs sequenced from human pre-frontal cortex30 were obtained from the Gene Expression Omnibus (GEO) database, accession numbers GSM450597-GSM450610. The sequencing reads were pre-processed using the mapper.pl script from the miRDeep2 package24 with the following options: -a -d -h -j -k TCGTATGCCG -l 18 -m. The RNA secondary structure of svtRNA2-1a and the sequencing read signature and the sequencing read counts across samples were computed using the quantifier.pl script from the same package, using default options. The sequencing read count of each sample was normalized by dividing by the total count of reads mapping to human miRNAs, also summed up with the quantifier.pl script. miRNAs were obtained from mirbase version 17.72 Two time points (34 d and 98 y) were sequenced in replicates, for these samples the mean normalized counts were used. Small RNAs sequenced from a Dicer-knockdown experiment were obtained from the GEO database accession numbers GSM769509-GSM769512. The expression fold-changes were calculated as previously described, except mirbase version 17 sequences were used and reads were mapped allowing a single mismatch.24
Cell culture and differentiation
SHSY-5Y cells were grown in DMEM medium supplemented with 10% of inactivated fetal bovine serum (FBS) and kept in low pass numbers (< 15). Standard differentiation protocol consisted of retinoic acid (RA) 10 µM exposure, followed by 12-O-tetradecanoylphorbol-13-acetate (TPA) 80 nM exposure, as previously described.20
Transient transfections
Differentiated SH-SY5Y cells were plated at day 3 of the differentiation process at 12,500 cells/mL and then transfected at day 10 using Lipofectamine 2000™ (Invitrogen) according to manufacturer’s instructions. The siRNAs and miRNA mimics used, as well as the concentrations used for transfection are listed in Table S6.
MTT cell viability assay
Cells were transfected in 96-well plates in 12 independent experiments; then at 24, 48 and 72 h post-transfection MTT was added to cell culture media at 0.5 mg/mL final concentration and incubated for 40 min at 37°C. Cells were lysed with 100 µL of DMSO upon medium removal and absorbance was measured at 550 nm. In each experiment, determinations were performed in quintuplicate. Percentage of cell viability was calculated using absorbance values from cells transfected with the anti-scrambled sequence as a reference. Statistical differences among the svtRNA2-1a- and the scrambled-transfected cells were determined using a t-test.
Supplementary Material
Acknowledgments
We thank G. Meister for providing the Ago2 antibodies for IP experiments. This work was supported by the Spanish Government: PN de I+D+I 2008-2011, PI081367 and PN de I+D+I 2012-2015 PI11/02036, Instituto Carlos III ISCIII, Subdirección General de Evaluación y Fomento de la Investigación (E.M.), SAF2008-00357 Ministerio de Economia y competitividad, ISCIII (X.E.), Centro de Investigación Biomédica en Red en Epidemiología y Salud Pública (CIBERESP) through the ISCIII (X.E., E.M., E.M.-M., B.K., G.E., M.F.); Centro de Investigación Biomédica en Red sobre Enfermedades Neurodegenerativas (CIBERNED) through the ISCIII” (I.F., S.P.); the Sixth Framework Programme of the European Commission through the SIROCCO integrated project LSHG-CT-2006-037900. The Spanish Government supports: E.M.-M. (BEFI contract, ISCIII), S.P. (Sara Borrell Postdoctoral contrat) and partially supports E.M. (Programa Miguel Servet). Support for M.R.F. was provided by EMBO long-term fellowship ALTF 225-2011.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24813
References
- 1.Braak H, Ghebremedhin E, Rüb U, Bratzke H, Del Tredici K. Stages in the development of Parkinson’s disease-related pathology. Cell Tissue Res. 2004;318:121–34. doi: 10.1007/s00441-004-0956-9. [DOI] [PubMed] [Google Scholar]
- 2.Cheng HC, Ulane CM, Burke RE. Clinical progression in Parkinson disease and the neurobiology of axons. Ann Neurol. 2010;67:715–25. doi: 10.1002/ana.21995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shulman JM, De Jager PL, Feany MB. Parkinson’s disease: genetics and pathogenesis. Annu Rev Pathol. 2011;6:193–222. doi: 10.1146/annurev-pathol-011110-130242. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Oroz MC, Jahanshahi M, Krack P, Litvan I, Macias R, Bezard E, et al. Initial clinical manifestations of Parkinson’s disease: features and pathophysiological mechanisms. Lancet Neurol. 2009;8:1128–39. doi: 10.1016/S1474-4422(09)70293-5. [DOI] [PubMed] [Google Scholar]
- 5.Braak H, Del Tredici K, Rüb U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging. 2003;24:197–211. doi: 10.1016/S0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 6.Douglas MR, Lewthwaite AJ, Nicholl DJ. Genetics of Parkinson’s disease and parkinsonism. Expert Rev Neurother. 2007;7:657–66. doi: 10.1586/14737175.7.6.657. [DOI] [PubMed] [Google Scholar]
- 7.Tan EK, Skipper LM. Pathogenic mutations in Parkinson disease. Hum Mutat. 2007;28:641–53. doi: 10.1002/humu.20507. [DOI] [PubMed] [Google Scholar]
- 8.Xiromerisiou G, Dardiotis E, Tsimourtou V, Kountra PM, Paterakis KN, Kapsalaki EZ, et al. Genetic basis of Parkinson disease. Neurosurg Focus. 2010;28:E7. doi: 10.3171/2009.10.FOCUS09220. [DOI] [PubMed] [Google Scholar]
- 9.Eriksen JL, Przedborski S, Petrucelli L. Gene dosage and pathogenesis of Parkinson’s disease. Trends Mol Med. 2005;11:91–6. doi: 10.1016/j.molmed.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Grünblatt E, Mandel S, Jacob-Hirsch J, Zeligson S, Amariglo N, Rechavi G, et al. Gene expression profiling of parkinsonian substantia nigra pars compacta; alterations in ubiquitin-proteasome, heat shock protein, iron and oxidative stress regulated proteins, cell adhesion/cellular matrix and vesicle trafficking genes. J Neural Transm. 2004;111:1543–73. doi: 10.1007/s00702-004-0212-1. [DOI] [PubMed] [Google Scholar]
- 11.Mandel S, Grunblatt E, Riederer P, Amariglio N, Jacob-Hirsch J, Rechavi G, et al. Gene expression profiling of sporadic Parkinson’s disease substantia nigra pars compacta reveals impairment of ubiquitin-proteasome subunits, SKP1A, aldehyde dehydrogenase, and chaperone HSC-70. Ann N Y Acad Sci. 2005;1053:356–75. doi: 10.1196/annals.1344.031. [DOI] [PubMed] [Google Scholar]
- 12.Simunovic F, Yi M, Wang Y, Macey L, Brown LT, Krichevsky AM, et al. Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson’s disease pathology. Brain. 2009;132:1795–809. doi: 10.1093/brain/awn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, James M, Middleton FA, Davis RL. Transcriptional analysis of multiple brain regions in Parkinson’s disease supports the involvement of specific protein processing, energy metabolism, and signaling pathways, and suggests novel disease mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2005;137B:5–16. doi: 10.1002/ajmg.b.30195. [DOI] [PubMed] [Google Scholar]
- 14.Büeler H. Impaired mitochondrial dynamics and function in the pathogenesis of Parkinson’s disease. Exp Neurol. 2009;218:235–46. doi: 10.1016/j.expneurol.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Cohen G. Oxidative stress, mitochondrial respiration, and Parkinson’s disease. Ann N Y Acad Sci. 2000;899:112–20. doi: 10.1111/j.1749-6632.2000.tb06180.x. [DOI] [PubMed] [Google Scholar]
- 16.Navarro A, Boveris A, Bández MJ, Sánchez-Pino MJ, Gómez C, Muntané G, et al. Human brain cortex: mitochondrial oxidative damage and adaptive response in Parkinson disease and in dementia with Lewy bodies. Free Radic Biol Med. 2009;46:1574–80. doi: 10.1016/j.freeradbiomed.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–4. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G, van der Walt JM, Mayhew G, Li YJ, Züchner S, Scott WK, et al. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82:283–9. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A. 2009;106:13052–7. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miñones-Moyano E, Porta S, Escaramís G, Rabionet R, Iraola S, Kagerbauer B, et al. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet. 2011;20:3067–78. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- 21.Persson H, Kvist A, Vallon-Christersson J, Medstrand P, Borg A, Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol. 2009;11:1268–71. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 22.Jellinger KA. Lewy body-related alpha-synucleinopathy in the aged human brain. J Neural Transm. 2004;111:1219–35. doi: 10.1007/s00702-004-0138-7. [DOI] [PubMed] [Google Scholar]
- 23.Saito Y, Ruberu NN, Sawabe M, Arai T, Kazama H, Hosoi T, et al. Lewy body-related alpha-synucleinopathy in aging. J Neuropathol Exp Neurol. 2004;63:742–9. doi: 10.1093/jnen/63.7.742. [DOI] [PubMed] [Google Scholar]
- 24.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Suzuki H, Hayashizaki Y, et al. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8:158–77. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–47. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 28.Sutherland GT, Matigian NA, Chalk AM, Anderson MJ, Silburn PA, Mackay-Sim A, et al. A cross-study transcriptional analysis of Parkinson’s disease. PLoS One. 2009;4:e4955. doi: 10.1371/journal.pone.0004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taccioli C, Tegnér J, Maselli V, Gomez-Cabrero D, Altobelli G, Emmett W, et al. ParkDB: a Parkinson’s disease gene expression database. Database (Oxford) 2011;2011:bar007. doi: 10.1093/database/bar007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Somel M, Guo S, Fu N, Yan Z, Hu HY, Xu Y, et al. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 2010;20:1207–18. doi: 10.1101/gr.106849.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vistica DT, Skehan P, Scudiero D, Monks A, Pittman A, Boyd MR. Tetrazolium-based assays for cellular viability: a critical examination of selected parameters affecting formazan production. Cancer Res. 1991;51:2515–20. [PubMed] [Google Scholar]
- 32.Berridge MV, Horsfield JA, Tan AS. Evidence that cell survival is controlled by interleukin-3 independently of cell proliferation. J Cell Physiol. 1995;163:466–76. doi: 10.1002/jcp.1041630306. [DOI] [PubMed] [Google Scholar]
- 33.Enciu AM, Popescu BO, Gheorghisan-Galateanu A. MicroRNAs in brain development and degeneration. Mol Biol Rep. 2012;39:2243–52. doi: 10.1007/s11033-011-0973-1. [DOI] [PubMed] [Google Scholar]
- 34.Hébert SS, Horré K, Nicolaï L, Bergmans B, Papadopoulou AS, Delacourte A, et al. MicroRNA regulation of Alzheimer’s Amyloid precursor protein expression. Neurobiol Dis. 2009;33:422–8. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 35.Hébert SS, Horré K, Nicolaï L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–20. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson R, Buckley NJ. Gene dysregulation in Huntington’s disease: REST, microRNAs and beyond. Neuromolecular Med. 2009;11:183–99. doi: 10.1007/s12017-009-8063-4. [DOI] [PubMed] [Google Scholar]
- 37.Li YY, Cui JG, Hill JM, Bhattacharjee S, Zhao Y, Lukiw WJ. Increased expression of miRNA-146a in Alzheimer’s disease transgenic mouse models. Neurosci Lett. 2011;487:94–8. doi: 10.1016/j.neulet.2010.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, et al. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A. 2010;107:15163–8. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nandy C, Mrázek J, Stoiber H, Grässer FA, Hüttenhofer A, Polacek N. Epstein-barr virus-induced expression of a novel human vault RNA. J Mol Biol. 2009;388:776–84. doi: 10.1016/j.jmb.2009.03.031. [DOI] [PubMed] [Google Scholar]
- 41.Stadler PF, Chen JJ, Hackermüller J, Hoffmann S, Horn F, Khaitovich P, et al. Evolution of vault RNAs. Mol Biol Evol. 2009;26:1975–91. doi: 10.1093/molbev/msp112. [DOI] [PubMed] [Google Scholar]
- 42.Berger W, Steiner E, Grusch M, Elbling L, Micksche M. Vaults and the major vault protein: novel roles in signal pathway regulation and immunity. Cell Mol Life Sci. 2009;66:43–61. doi: 10.1007/s00018-008-8364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Zon A, Mossink MH, Scheper RJ, Sonneveld P, Wiemer EA. The vault complex. Cell Mol Life Sci. 2003;60:1828–37. doi: 10.1007/s00018-003-3030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee K, Kunkeaw N, Jeon SH, Lee I, Johnson BH, Kang GY, et al. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA. 2011;17:1076–89. doi: 10.1261/rna.2701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–6. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–6. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pillai MM, Yang X, Balakrishnan I, Bemis L, Torok-Storb B. MiR-886-3p down regulates CXCL12 (SDF1) expression in human marrow stromal cells. PLoS One. 2010;5:e14304. doi: 10.1371/journal.pone.0014304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asaoka T, Sotolongo B, Island ER, Tryphonopoulos P, Selvaggi G, Moon J, et al. MicroRNA signature of intestinal acute cellular rejection in formalin-fixed paraffin-embedded mucosal biopsies. Am J Transplant. 2012;12:458–68. doi: 10.1111/j.1600-6143.2011.03807.x. [DOI] [PubMed] [Google Scholar]
- 50.Li JH, Xiao X, Zhang YN, Wang YM, Feng LM, Wu YM, et al. MicroRNA miR-886-5p inhibits apoptosis by down-regulating Bax expression in human cervical carcinoma cells. Gynecol Oncol. 2011;120:145–51. doi: 10.1016/j.ygyno.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 51.Xiong Y, Zhang L, Holloway AK, Wu X, Su L, Kebebew E. MiR-886-3p regulates cell proliferation and migration, and is dysregulated in familial non-medullary thyroid cancer. PLoS One. 2011;6:e24717. doi: 10.1371/journal.pone.0024717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bañez-Coronel M, Porta S, Kagerbauer B, Mateu-Huertas E, Pantano L, Ferrer I, et al. A pathogenic mechanism in Huntington’s disease involves small CAG-repeated RNAs with neurotoxic activity. PLoS Genet. 2012;8:e1002481. doi: 10.1371/journal.pgen.1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dexter DT, Sian J, Rose S, Hindmarsh JG, Mann VM, Cooper JM, et al. Indices of oxidative stress and mitochondrial function in individuals with incidental Lewy body disease. Ann Neurol. 1994;35:38–44. doi: 10.1002/ana.410350107. [DOI] [PubMed] [Google Scholar]
- 54.Ferrer I. Early involvement of the cerebral cortex in Parkinson’s disease: convergence of multiple metabolic defects. Prog Neurobiol. 2009;88:89–103. doi: 10.1016/j.pneurobio.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 55.Ferrer I, Martinez A, Blanco R, Dalfó E, Carmona M. Neuropathology of sporadic Parkinson disease before the appearance of parkinsonism: preclinical Parkinson disease. J Neural Transm. 2011;118:821–39. doi: 10.1007/s00702-010-0482-8. [DOI] [PubMed] [Google Scholar]
- 56.Parker WD, Jr., Parks JK, Swerdlow RH. Complex I deficiency in Parkinson’s disease frontal cortex. Brain Res. 2008;1189:215–8. doi: 10.1016/j.brainres.2007.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schulz-Schaeffer WJ. The synaptic pathology of alpha-synuclein aggregation in dementia with Lewy bodies, Parkinson’s disease and Parkinson’s disease dementia. Acta Neuropathol. 2010;120:131–43. doi: 10.1007/s00401-010-0711-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanji K, Mori F, Mimura J, Itoh K, Kakita A, Takahashi H, et al. Proteinase K-resistant alpha-synuclein is deposited in presynapses in human Lewy body disease and A53T alpha-synuclein transgenic mice. Acta Neuropathol. 2010;120:145–54. doi: 10.1007/s00401-010-0676-z. [DOI] [PubMed] [Google Scholar]
- 59.Mrázek J, Kreutmayer SB, Grässer FA, Polacek N, Hüttenhofer A. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res. 2007;35:e73. doi: 10.1093/nar/gkm244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–7. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 62.Bohnen NI, Koeppe RA, Minoshima S, Giordani B, Albin RL, Frey KA, et al. Cerebral glucose metabolic features of Parkinson disease and incident dementia: longitudinal study. J Nucl Med. 2011;52:848–55. doi: 10.2967/jnumed.111.089946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bohnen NI, Minoshima S, Giordani B, Frey KA, Kuhl DE. Motor correlates of occipital glucose hypometabolism in Parkinson’s disease without dementia. Neurology. 1999;52:541–6. doi: 10.1212/WNL.52.3.541. [DOI] [PubMed] [Google Scholar]
- 64.Eberling JL, Richardson BC, Reed BR, Wolfe N, Jagust WJ. Cortical glucose metabolism in Parkinson’s disease without dementia. Neurobiol Aging. 1994;15:329–35. doi: 10.1016/0197-4580(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 65.Otsuka M, Ichiya Y, Kuwabara Y, Hosokawa S, Sasaki M, Yoshida T, et al. Glucose metabolism in the cortical and subcortical brain structures in multiple system atrophy and Parkinson’s disease: a positron emission tomographic study. J Neurol Sci. 1996;144:77–83. doi: 10.1016/S0022-510X(96)00172-4. [DOI] [PubMed] [Google Scholar]
- 66.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 67.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 68.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57:289–300. [Google Scholar]
- 71.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.