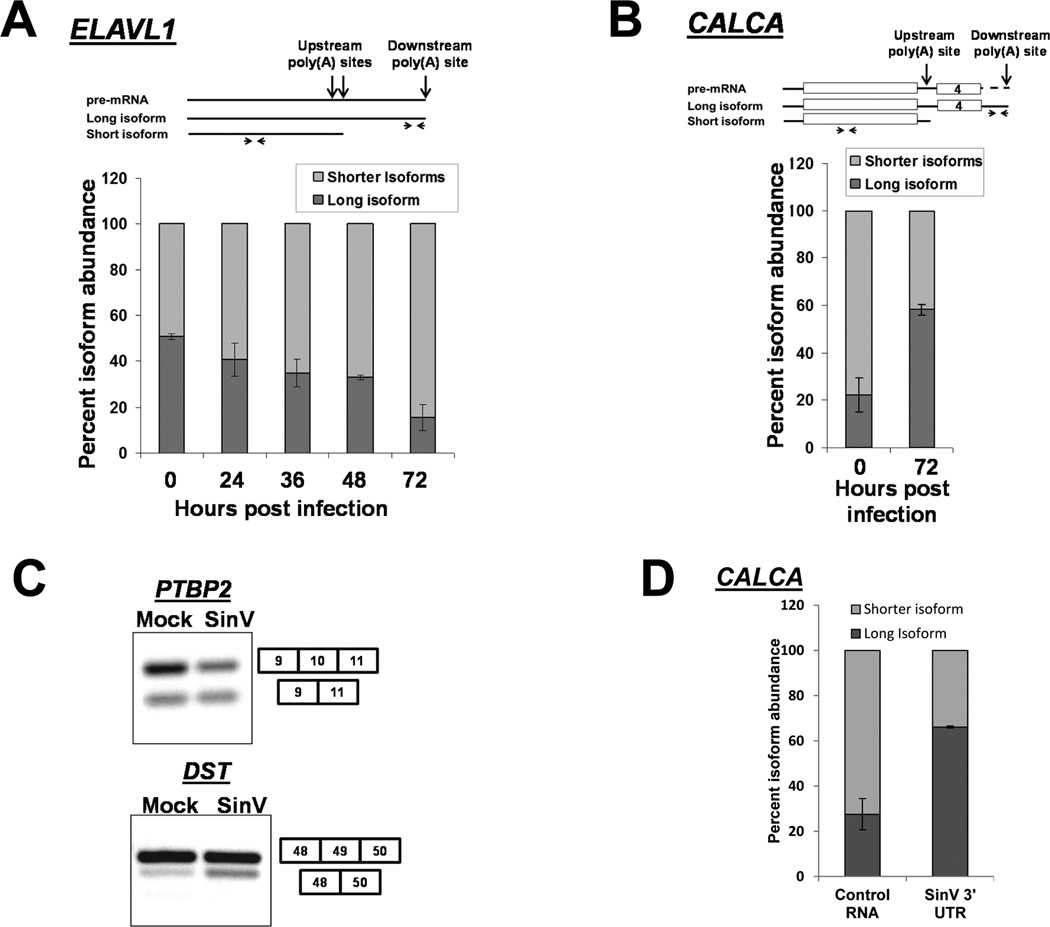
Figure 4. SinV infection influences alternative polyadenylation and splicing of cellular pre-mRNAs.
293T cells were infected with SinV for the indicated times. Short or long isoforms of the ELAVL1 (HuR) mRNA formed by alternative polyadenylation (Panel A) or the indicated isoforms of CALCA (calcitonin) were quantified by qRT-PCR using the primers illustrated at the top of the panels. Note that the level of ‘shorter isoforms’ was determined by subtraction of the amount of the longer isoform from the level of total RNA detected by the upstream primer. Quantification is shown with standard deviations calculated from three independent experiments. Panel C. Total RNA was isolated 24 hpi, probed for the presence of the splicing isoforms of the indicated genes by RT-PCR and analyzed on a 2% agarose gel. Panel D: 293T cells were transfected with equimolar amounts of either a control RNA (GemA60) or the SinV 3’UTR RNA. Total RNA was isolated at 6 hours post transfection and analyzed for CALCA isoforms as described in Panel B above. Quantification is shown with standard deviations calculated from two independent experiments.