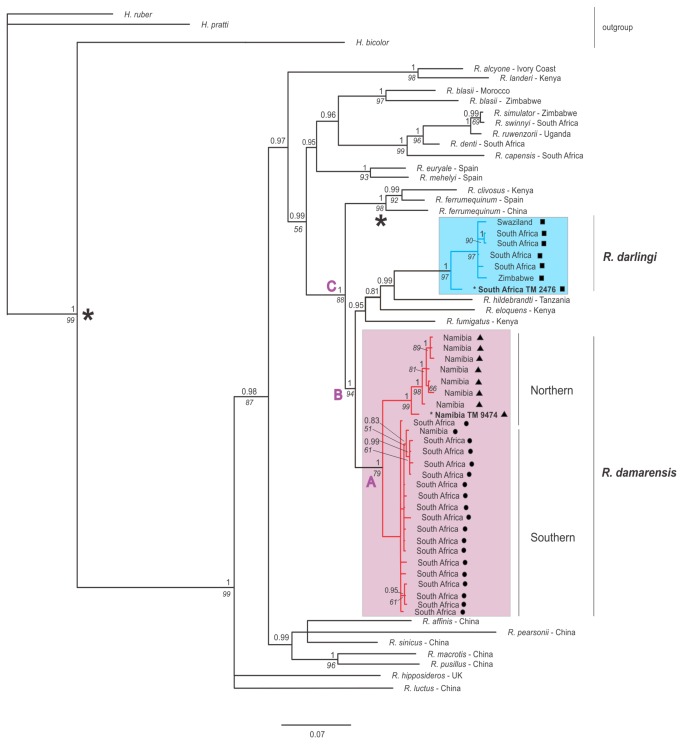
Figure 2. Bayesian consensus topology of Rhinolophus spp. based on cytochrome b.
Values above nodes indicate Bayesian posterior probabilities (> 0.80) and below nodes report parsimony bootstrap support. Branch lengths are proportional to average number of substitutions per site. Divergence times (millions years ago, Mya) were estimated for three nodes of interest: node A = 4.8 Mya (95% HPD; 2.69–7.52), B = 9.68 Mya (95% HPD; 5.87–10.04), C = 10.51 Mya (95% HPD; 6.65–15.43). The split between the Hipposideridae (outgroup) and the Rhinolophidae together with fossil dates for R. ferrumequinum were used as calibration points; calibration points are indicated on the tree with an asterisk *. Sequences from the type localities for R. darlingi darlingi (TM2476) and R. darlingi damarensis (TM9474) are indicated on the tree in bold. The eastern R. darlingi darlingi subspecies forms a single clade together with R. fumigatus, R. eloquens and R. hildebrandti (blue) while R. damarensis damarensis, distributed in the western region of southern Africa, is composed of two very well supported clades that subdivide into northern and southern lineages (red) that diverged in the late Miocene.