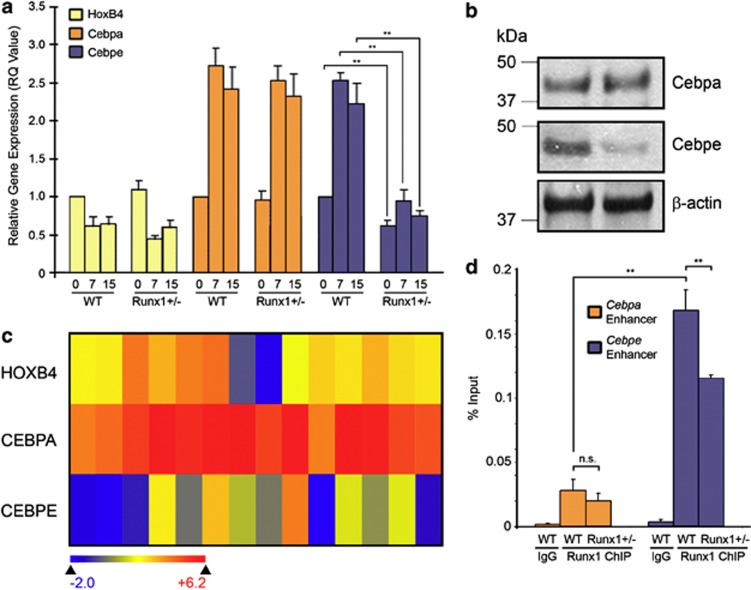
Figure 2.
Unequal impact of Runx1 deficiency on Runx1 binding at the Cebpa and Cebpe enhancers and on Cebpa and Cebpe activation in response to G-CSF. (a) Time-course expression of Hoxb4 (key stem cell transcription factor), Cebpa (key lineage-commitment transcription factor) and Cebpe (key late-differentiation transcription factor) in WT and Runx1+/− lineage-negative (Lin−) cells cultured with G-CSF for 15 days. SYBR Green bound to double-stranded DNA was detected in real-time with the 7500 Fast Real Time PCR System (Applied Biosystems, Foster City, CA, USA). Relative expression values were calculated by raising 2 to the power of the negative value of delta-delta CT for each sample. Primer sequences: HoxB4: forward (F): 5′-CCAGAATCGGCGCATGA-3, reverse (R): 5′-CCCGAGCGGATCTTGGT-3′ Cebpa: F: 5′-CAAAGCCAAGAAGTCGGTGGACAA-3′ R: 5′-TCATTGTCACTGGTCAACTCCAGC-3′ Cebpe: F: 5′-GCTACAATCCCCTGCAGTACC-3′ R: 5′-TGCCTTCTTGCCCTTGTG-3′ GAPDH: F: 5′-ACCACACTCCATGCCATCAC-3′, R:5′-TCCACCACCCTGTTGCTGTA-3′. Mean±standard deviation. **P<0.01, Student's t-test. Experiments performed in triplicate. (b) Cebpa and Cebpe protein expression in WT and Runx1+/− Lin− cells cultured for 15 days with G-CSF. Western blot analyses with the following primary antibodies: anti-Cebpa, anti-Cebpe (Santa Cruz Biotechnology, Dallas, TX, USA; #SC-61, #SC-25770), anti-β-actin (Sigma-Aldrich, St Louis, MO, USA, A3854); secondary antibodies: anti-Rabbit (GE Healthcare, Waukesha, WI, USA; NA934) and anti-Mouse (GE Healthcare, NXA931) at 1:5000 and 1:10 000 dilutions, respectively. (c) Gene expression of CEBPA, CEBPE and HOXB4 in primary AML cells with normal cytogenetics and RUNX1 mutation (TCGA database, n=14). Heatmap by Arraystar software. (d) Chromatin immunoprecipitation (ChIP) analysis of Runx1+/− and WT cells to evaluate Runx1 binding at the Cepba and Cebpe enhancers. Enhancers of Cebpa and Cebpe were identified by others using ChIP coupled with deep sequencing.28 ChIP performed using ChIP Assay Kit (cat#17-295, EMD Millipore, Billerica, MA, USA). 5 × 106 cells bone marrow (BM) cells from WT and Runx1+/1 fresh BM cells were resuspended in 10 ml of media and formaldehyde was added to a final concentration of 1% followed by 10 min incubation at 37 °C. The media containing formaldehyde was removed and the cells were washed twice with ice-cold PBS containing protease inhibitor (cat#P8340, Sigma-Aldrich) and phosphatase inhibitor cocktails (cat#P0044, Sigma-Aldrich). Crosslinked cells were resuspended in SDS lysis buffer (Millipore, #20–163) and incubated for 10 min on ice. These cell pellets were sonicated at 15 s pulses with 45 s hold on ice for a total of 10 min of pulsing at full power (Fisher-Scientific Model #550 Sonic Dismembrator equipped with a microtip probe). Genomic DNA fragments of 200-bp and 1-kb in size were obtained (confirmed by agarose gel electrophoresis). After centrifugation at 4 °C, the supernatant was pre-cleared with salmon sperm DNA/Protein A agarose-50% slurry (Millipore, #16–157) for 30 min at 4 °C with agitation. A 10% aliquot (vol) of the precleared protein/DNA mixture was removed and used for subsequent reverse transcription–PCR input quantification. 10 μg of anti-Runx1 ChIP grade antibody (cat#ab23980, Abcam, Cambridge, MA, USA) was added to the remaining protein/DNA mixture and incubated at 4 °C overnight with rotation. Salmon sperm DNA/Protein A agarose-50% slurry was added and incubated for 1 h at 4 °C followed by washes with the ChIP assay kit provided buffers. The immunoprecipitated (IP) complexes were reverse crosslinked at 65 °C for 4 h and DNA was recovered by phenol/chloroform extraction protocol (Sigma). The IP products were amplified and quantified using real-time PCR. ChIP primer sequences are:21 cepba enhancer left primer, 5′- TTCCCGTTTCTGAAATCTGC-3′, cebpa enhancer right primer, 5′-GGTTGTGGCAAGAAGGTCAC-3′, cebpe enhancer left primer, 5′-GTGTCATGGTCACCCTAGCC-3′ and cebpe enhancer right primer, 5′-CTGGAGCTAGCAGGGGTTTT-3′. Mean±standard deviation from three independent ChIP experiments. **P<0.001, NS=not significant, Tukey–Kramer HSD test.