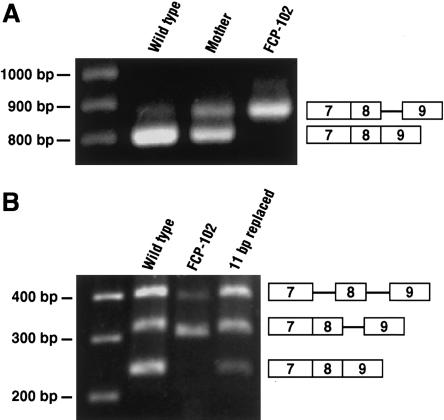
Figure 2.
RT-PCR showing both splicing error due to intronic deletion and correction of defect by restoration of intron size. A, RT-PCR of lymphoblast RNA from proband FCP-102, his mother, and normal control. Primers are within exons 5 and 11 (between the white arrows in fig. 1A). The normal product is expected to be 791 bp. The aberrant larger product is 857 bp. B, RT-PCR of transcripts from NIH3T3 cells transfected with expression constructs containing exons 5–10 of RECQL4 from wild type, FCP-102, or wild type replaced with 11 unrelated base pairs at the deletion site. Primers are within exons 7 and 9 (between the black arrows in fig. 1A). Predicted sizes are 240 bp, for correct splicing of intron 8; 306 bp, for retention of intron 8 containing the deletion; and 317 bp, for the retention of wild-type intron 8. The highest band in each lane represents retention of introns 7 and 8 (389 bp for FCP-102 and 400 bp for wild type and the replaced construct).