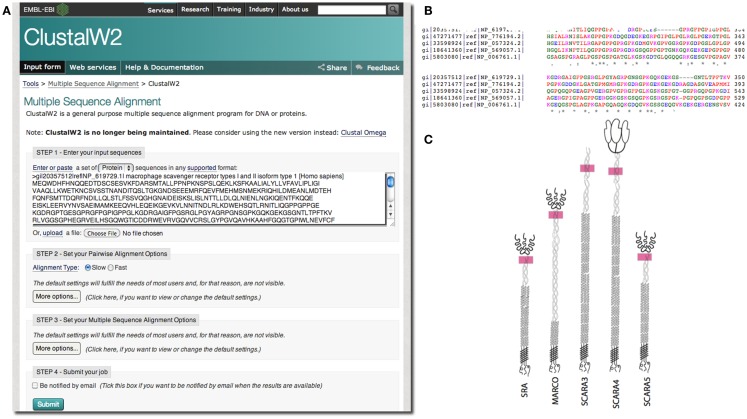
Figure 3.
Use of multiple sequence alignments to discover regions of evolutionary conservation and presumed functionality. FASTA formatted protein sequences of the scavenger receptors were obtained as described previously for SRAI (NP_619729.1), MARCO (NP_006761.1), SCARA3 (NP_057324.2), SCARA4 (NP_569057.1), and SCARA5 (NP_776194.2) and inputted into the Multiple Sequence Alignment tool, ClustalW2 (A). The sequences were aligned; a portion of the alignment with the highest conservation across all five sequences is shown (B). The user may choose to view colored output, where red represents small, hydrophobic amino acids (AVFPMILW), blue represents acidic amino acids (DE), magenta represents basic amino acids (RK), and green represents STYHCNGQ (hydroxyl, sulfhydryl, amine, and glycine). Coloring allows the viewer to visualize the distribution of charge and hydrophobicity in the protein. In this example, we see that there is an orderly distribution of hydrophobic amino acids (red). The degree of consensus is represented with symbols. (*) Indicates positions which have a single, fully conserved residue; (:) indicates conservation between groups of strongly similar properties; (.) represents conservation between groups of amino acids with weakly similar properties. The fact that all five members of this family share this highly conserved region at locations in these proteins indicated with pink rectangles, (C), and that it is the highest area of conservation within the proteins is strongly suggestive of a conserved function.