Figure 2.
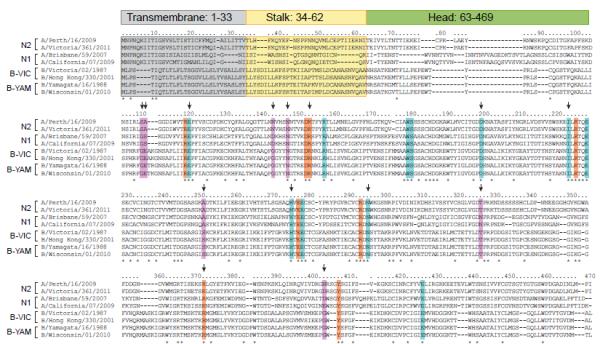
Alignment of NA proteins of N1 and N2 subtypes of influenza A viruses and influenza B viruses. Transmembrane, stalk, and head regions of NA protein are indicated by gray, yellow, or green shading, respectively. Catalytic residues are highlighted in orange (R118, D151, R152, R224, E276, R292, R371, and Y406), framework residues in teal (E119, R156, W178, S179, D198, I222, E227, H274, E277, N294, and E425). Residues associated with NAI resistance in influenza B viruses from clinical, surveillance, or experimental studies are designated with an arrow, with other supporting residues (not catalytic or framework) shown in lavender (G109, E110, S250, T325, G402, G142+N146). Strain subtype and years of use in vaccine composition (shown in parentheses): A/Perth/16/2009 (H3N2, 2010-2012); A/Victoria/361/2011 (H3N2); A/Brisbane/59/2007 (H1N1, 2008-2010); A/California/07/2009 (H1N1pdm, 2010-2013); B/Victoria/02/1987 (HA and NA genes from Victoria lineage); B/Hong Kong/330/2001 (HA and NA genes from Victoria lineage, 2002-2004); B/Yamagata/16/1988 (HA and NA genes from Yamagata lineage, 1989-1992); B/Wisconsin/01/2010 (HA and NA genes from Yamagata lineage, 2012-2013).
