Fig. 7.
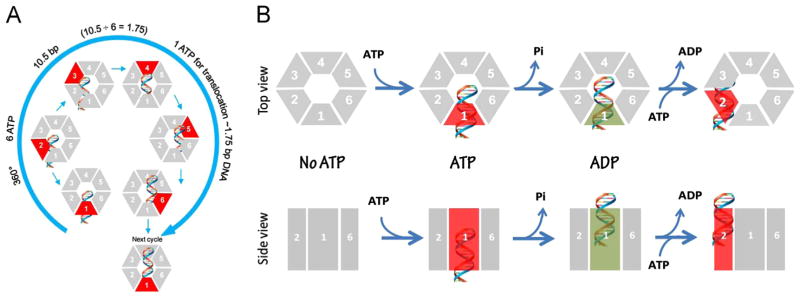
Schematic of gp16 binding to DNA and mechanism of sequential revolution in translocating genomic DNA. The connector is a one way valve that allows dsDNA to move into the procapsid, but does not allow movement in the opposite direction. Gp16, which is bridged by pRNA to associate with the connector, provides the pushing force. The binding of ATP to one subunit stimulates gp16 to adopt a conformation with a higher affinity for dsDNA. ATP hydrolysis forces gp16 to assume a new conformation with a lower affinity for dsDNA, thus pushing dsDNA away from the subunit and transferring it to an adjacent subunit. DsDNA is translocated at a pace of 1.75 base pairs per transfer to the neighboring subunit and is bound at a location 60° different from the first subunit on the same phosphate backbone chain. Rotation of neither the hexameric ring nor the dsDNA is required since the dsDNA revolves around the diameter of the ATPase. In each transitional step, one ATP is hydrolyzed, and in one cycle, six ATPs are required to translocate dsDNA one helical turn of 360° (10.5 base pairs). An animation is available at http://nanobio.uky.edu/movie.html.
