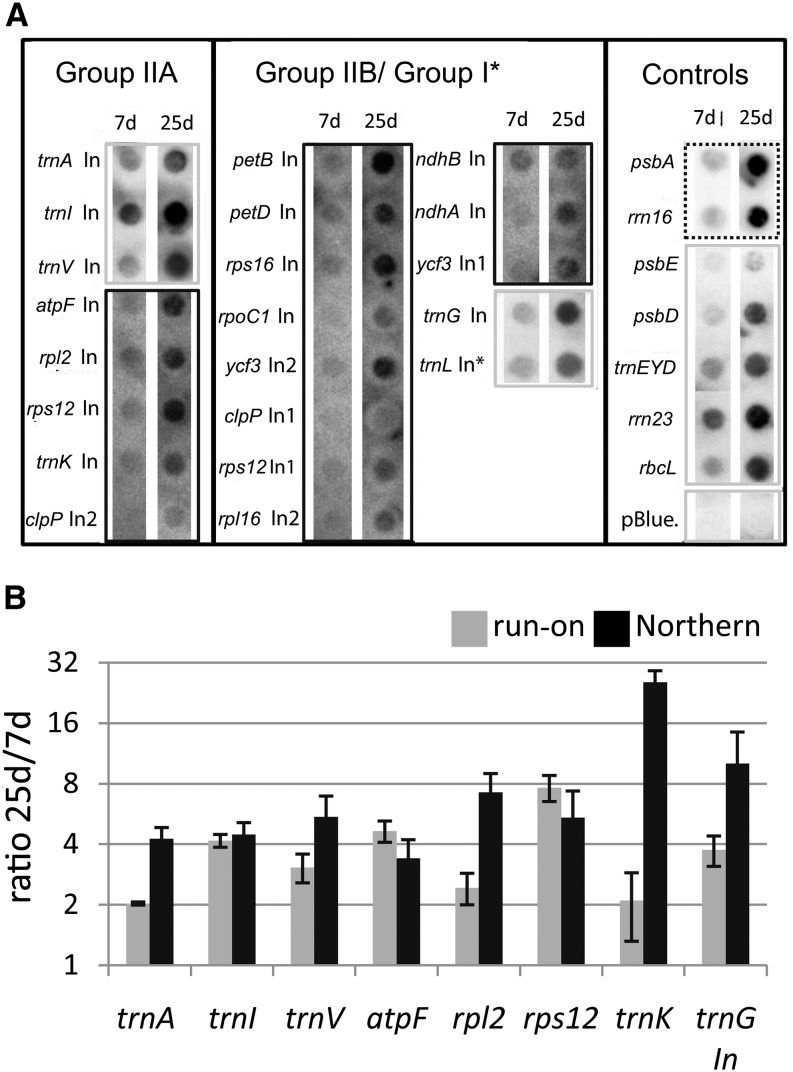
Figure 2.
Run-on transcription analysis of all intron-containing genes in the chloroplast and additional controls. A, RNA was extracted from run-on reactions, which were carried out with chloroplasts purified from 7- and 25-d-old plants and hybridized on macroarrays. The array comprised probes for all chloroplast introns (In) plus additional controls, including a negative control (pBlue., pBluescript II SK+). Probes were spotted as duplicates (only one spot shown). The signals shown here are examples of two biological replicates. The data were used to generate the charts in B. For better visualization, different exposure times are shown for different probes (dashed line = short; gray lines = intermediate; black lines = long). B, Comparison of transcription rates and RNA steady-state levels in 25- versus 7-d-old material. mRNA levels were assayed by northern blot and quantified from two independent experiments (gray bars). Transcription rates were assayed by run-on analysis and quantified from two independent assays (black bars; see A). Note the logarithmic scale of the y axis. Assayed genes are indicated below. sd values were derived from two biological replicates.