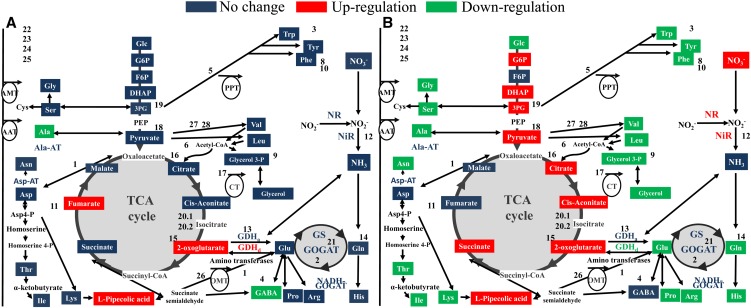
Figure 4.
The responses of C and N metabolism in wild-type and transgenic PSARK::IPT rice plants to water stress. A, Changes in C and N metabolism in transgenic PSARK::IPT rice plants relative to wild-type plants grown under well-watered conditions. B, Changes in C and N metabolism in transgenic PSARK::IPT rice plants relative to wild-type plants grown under water-stress conditions. Metabolites are represented in color boxes and include amino acids (Ala, Arg, Asn, Asp, Glu, Gln, Gly, His, Ile, Leu, Lys, l-pipecolic acid, Met, Phe, Pro, Ser, Thr, Trp, Tyr, and Val), TCA- and glycolysis-related compounds (Glc, Glc-6-P [G6P], Fru-6-P [F6P], dihydroxyacetone phosphate [DHAP], pyruvate, citrate, cis-aconitate, 2-oxoglutarate, succinate, fumarate, malate, glycerol 3-phosphate, and glycerol), and N assimilation compounds (NO3−, NO2−, and NH4+). Assayed enzymes are presented in boldface (NR, NiR, GS, NADH-GOGAT, GDH amination [GDHa] and deamination [GDHd] directions, Ala aminotransferase [Ala-AT], and Asp aminotransferase [Asp-AT]). Numbers represent genes that are differently expressed (listed in Supplemental Fig. S1): 1 (LOC_Os02g14110, aminotransferase), 1.2 (LOC_Os01g08270, aminotransferase), 1.3 (LOC_Os09g28050, aminotransferase), 1.4 (LOC_Os03g18810, aminotransferase), 1.5 (LOC_Os01g55540, aminotransferase), 2 (LOC_Os07g46460, Fd-GOGAT), 3 (LOC_Os02g20360, Tyr aminotransferase), 4 (LOC_Os04g52450, aminotransferase), 5 (LOC_Os08g01410, phosphate/phosphoenolpyruvate translocator), 6 (LOC_Os01g02020, acetyl-CoA acetyltransferase), 7 (LOC_Os07g42600, Ala aminotransferase), 8 (LOC_Os02g07160, glyoxalase family protein), 9 (LOC_Os04g14790, glycerol-3-phosphate dehydrogenase), 10 (LOC_Os02g41680, Phe ammonia-lyase), 11 (LOC_Os04g33300, amino acid kinase), 12 (LOC_Os02g37040, NTP3 [nitrate transporter]), 13 (LOC_Os04g45970, GDH), 14 (LOC_Os01g09000, glutaminyl-tRNA synthetase), 14.2 (LOC_Os07g07260, Gln-dependent NAD), 15 (LOC_Os02g38200, isocitrate dehydrogenase), 15.2 (LOC_Os01g46610, isocitrate dehydrogenase), 16 (LOC_Os02g10070, citrate synthase), 17 (LOC_Os03g05390, citrate transporter), 18 (LOC_Os04g31700, methylisocitrate lyase 2), 19 (LOC_Os10g08550, enolase), 20 (LOC_Os08g09200, aconitate hydratase), 20.2 (LOC_Os03g04410, aconitate hydratase), 21 (LOC_Os04g56400, Gln synthetase), 22.1 (LOC_Os06g16420, amino acid transporter), 22.2 (LOC_Os04g12499, amino acid transporter), 22.3 (LOC_Os06g36210, amino acid transporter), 22.4 (LOC_Os06g43700, amino acid transporter), 22.4 (LOC_Os11g09020, amino acid transporter), 22.5 (LOC_Os12g42850, amino acid permease), 22.6 (LOC_Os03g25869, amino acid permease), 22.7 (LOC_Os08g23440, amino acid permease), 22.8 (LOC_Os10g30090, amino acid permease), 22.9 (LOC_Os11g05690, amino acid permease), 22.10 (LOC_Os04g35540, amino acid transporter), 22.11 (LOC_Os08g03350, amino acid transporter), 22.12 (LOC_Os01g40410, amino acid transporter), 22.13 (LOC_Os01g63854, amino acid transporter), 22.14 (LOC_Os02g44980, amino acid transporter), 22.15 (LOC_Os02g09810, amino acid transporter), 22.16 (LOC_Os04g38680, amino acid transporter), 22.17 (LOC_Os05g50920, amino acid transporter), 22.18 (LOC_Os01g65000, ammonium transporter protein), 23.1 (LOC_Os02g34580, ammonium transporter protein), 23.2 (LOC_Os03g62200, ammonium transporter protein), 23.3 (LOC_Os04g43070, ammonium transporter protein), 24.1 (LOC_Os03g13240, peptide transporter PTR2), 24.2 (LOC_Os06g49250, peptide transporter PTR2), 24.3 (LOC_Os10g02240, peptide transporter PTR2), 24.4 (LOC_Os10g33170, peptide transporter PTR2), 24.5 (LOC_Os03g51050, peptide transporter PTR2), 25.1 (LOC_Os03g13274, NO3 peptide transporter PTR2), 25.2 (LOC_Os10g42900, NO3 peptide transporter PTR3), 26 (LOC_Os11g24450, 2-oxoglutarate/malate translocator), 27 (LOC_Os08g43170, hydroxymethylglutaryl-CoA synthase), and 28 (LOC_Os02g32490, acetyl-CoA synthetase). Red, green, and blue colors indicate higher, lower, and unchanged metabolite content/enzymatic activity, respectively. One-way ANOVA followed by Student’s t test (P ≤ 0.05) was performed using the values (metabolite content, enzyme activity, or gene expression) from six different plants (n = 6) per genotype/treatment.