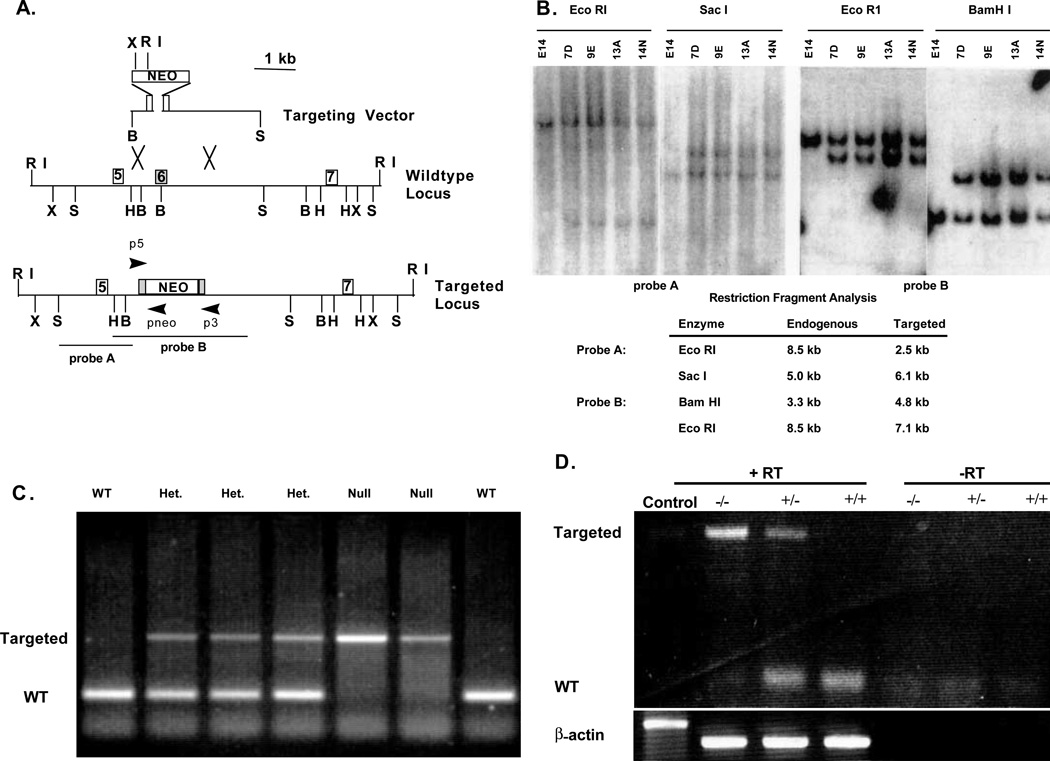
Fig. 1.
Targeting of the mouse TGFβ2 gene. (A) Diagrams of the wild-type locus, the targeting vector and the predicted targeted allele. Shown is the exon structure and the primers used for various PCR reactions to distinguish the targeted from the wild-type alleles in both ES cell and tail clip DNA. Abbreviation used: B, BamHI; E, EcoRI; H, HindIII; S, SacI; X, XbaI; neo, poly(A)– pMC1 neo cassette. (B) Southern blots of ES cell DNA showing both control cell DNA (E14) and targeted clones. DNA was digested with the enzymes indicated and probed with either probe A or probe B. Expected restriction fragments are listed. (C) PCR-genotype of the offspring from a heterozygote (HET) intercross. The null (1.3 kb) and the wild-type (132 bp) alleles are amplified using primers p3 and p5. (D) RT-PCR analysis from HET, wild-type and null animals showing the absence of detectable TGFβ2 exon 6-specific message in the null animals. +RT and –RT represent PCR substrates produced in the presence or absence of reverse transcriptase. The β-actin lanes are PCR controls for genomic DNA contamination. The β-actin control lane is genomic DNA only.