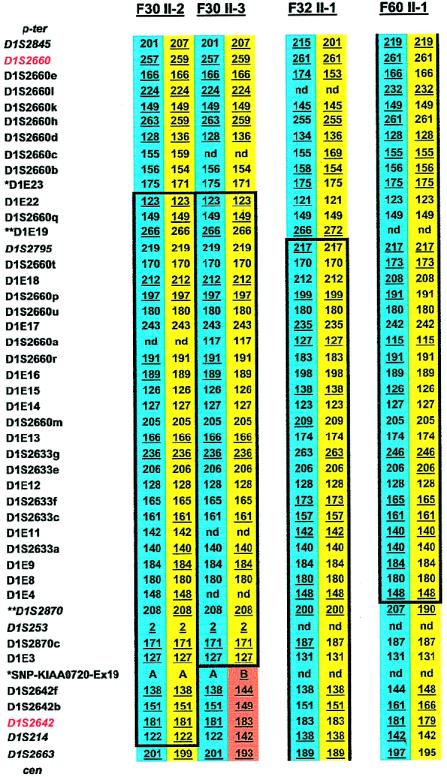
Figure 1.
Haplotype results on chromosome 1p36 performed for refinement of the NPHP4 locus in affected offspring from three consanguineous families with NPHP. Family, generation, and individual numbers are indicated above haplotypes. Paternal haplotypes are shown on blue background, and maternal haplotypes are shown on yellow background. A recombination in the maternal haplotype, which was directly observed in parent-to-child transmission in this pedigree, is shown on orange background. At left, 8 published microsatellites are given in italic, and 38 newly generated microsatellites are given in roman. Flanking markers to the published 2.1-Mb critical NPHP4 interval are depicted in red. Informative alleles are underlined. Haplotypes homozygous in continuity are encased in boxes. Homozygosity mapping revealed a p-terminal recombinant for marker D1E23 (asterisks) on the basis of heterozygosity in individuals F30 II-2 and F30 II-3 and an observed recombinant for marker SNP-KIAA0720-Ex19 (asterisks) in individual F30 II-3, thus refining the critical genetic region to a secure interval <1.2 Mb. On the basis of a significant LOD score yielded for F30 alone (Schuermann et al. 2002), this refinement yields secure borders. Further refinement was achieved through heterozygosity for markers D1E19 and D1S2870 (double asterisks) in individuals F32 II-1 and F60 II-1, respectively. This refines the critical genetic region to a suggestive interval <700 kb. Because of the presence of only two affected individuals in each family, this refinement is only suggestive. p-ter = Telomeric; cen = centromeric; nd = not done.