Abstract
In the past decade, a concerted effort to successfully capture specific tertiary packing interactions produced specific three-dimensional structures for many de novo designed proteins that are validated by nuclear magnetic resonance and/or X-ray crystallographic techniques. However, the success rate of computational design remains low. In this review, we provide an overview of experimentally validated, de novo designed proteins and compare four available programs, RosettaDesign, EGAD, Liang-Grishin, and RosettaDesign-SR, by assessing designed sequences computationally. Computational assessment includes the recovery of native sequences, the calculation of sizes of hydrophobic patches and total solvent-accessible surface area, and the prediction of structural properties such as intrinsic disorder, secondary structures, and three-dimensional structures. This computational assessment, together with a recent community-wide experiment in assessing scoring functions for interface design, suggests that the next-generation protein-design scoring function will come from the right balance of complementary interaction terms. Such balance may be found when more negative experimental data become available as part of a training set.
Keywords: computational assessment of designed sequences, secondary structure, solvent accessibility, hydrophobic patch, intrinsic disorder
INTRODUCTION
De novo protein design refers to computational design of new protein molecules that possess desired biological functions. Such computational design is needed to supplement and accelerate naturally occurring processes that can create conformationally and functionally novel proteins, as naturally occurring processes are constrained by biological functional requirements and limited by the tools available in nature. For example, one naturally occurring process that produces new topologically linked protein structures is circular permutation, a process that closes the N and C termini with a short loop and opens another loop for new termini (16, 71). This single-loop permutation, however, often results in essentially the same structure prior to circular permutation (42, 48). By comparison, new topologically folded structures can be generated more frequently in silico by changing the connections of multiple, rather than single, loops while maintaining the core packing (20). This and other studies (14, 111) suggest the existence of a vast, unexplored space of possible structural folds of proteins. A limited exploration of the protein structural space is more obvious for proteins with a knot in their polypeptide backbones. There are only 78 nonredundant knotted proteins in the entire Protein Data Bank of 30,000 structures (90% sequence identity cutoff), a number much lower than would be expected to occur by chance (46, 113). Most of these 78 knots are the simple three-point crossing (trefoil) knot, and the most complex is a six-point crossing knot for one protein called α-haloacid dehalogenase (the Stevedore knot) (55, 61). The rarity and simplicity of knotted proteins again suggest the opportunity to supplement natively knotted proteins with designed ones (20, 53). The functional space of proteins is also far from fully explored by nature. For example, enzymes can catalyze only a selected set of chemical reactions required for the life cycle of living organisms. Such vast unexplored structural and functional space of proteins has motivated active research in protein design, which is steadily increasing our knowledge of protein structure and function while more clearly defining opportunities for future explorations.
Significant strides in a number of areas have been made in the past two decades. In the early 1990s, most designed proteins had molten-globule-like structures with low stability (43, 52, 91, 92). Currently, on the other hand, specific structures of de novo designed proteins are routinely validated by NMR or X-ray structure determination (5, 9, 17, 21, 27, 41, 56, 60, 82, 104, 115). New structural folds were also successfully designed in 2003 (58) and 2009 (65). Progress in structural specificity and stability was accompanied by novel proteins designed with functions ranging from protein binding (3, 40, 54, 59, 74, 93, 105) to catalytic activities (7, 29, 50, 62, 84, 106) to conformational switches (1, 2). Such advances make it clear that de novo protein design holds promise to significantly accelerate the development of novel proteins for diagnostic, therapeutic, and industrial purposes.
This promise, however, is still unfulfilled largely because of the low success rate of de novo design (12, 34, 63, 72, 109). Dantas et al. (22) performed a large-scale test of nine proteins designed by RosettaDesign and found that only “half of the folded designs have NMR spectra and temperaturemelts typical of tightly packed proteins.” Schreier et al. (103) reexamined five computationally designed proteins and found that none of them performed as expected due to instability, aggregation, or lack of detectable designed ligand binding. Fleishman et al. (34) showed that only 2 of 73 designed proteins will bind with detectable binding affinity to the targeted stem region of influenza hemagglutinin.
To improve the success rate of de novo protein design, we must overcome two practical challenges. First, because experimentally measuring the success rate of designed proteins is time-consuming and costly, many studies have relied on manual inspection and human expertise in selecting designed sequences likely to be successful (72). As a result, we have yet to construct and test a fully automated design process that could offer an actual success rate high enough to warrant routine usage by biochemists. Second, because most protein design software is not openly available for academic users, few comparisons between different computational techniques have been made. These factors have made it difficult to determine what makes one design successful and another design unsuccessful.
To limit our scope, this review focuses on de novo design of protein structures. We compare four available protein design programs by computationally assessing designed sequences. We show how different balances of energetic terms lead to different outcomes in native sequence recovery, sizes of hydrophobic patches, and intrinsic disorder, among others. We propose that inaccurate scoring functions are the origin of low success rates of protein design. Locating the right balance for the right energy terms is the key to further improving protein design.
DE NOVO DESIGNED AND STRUCTURALLY VALIDATED PROTEINS
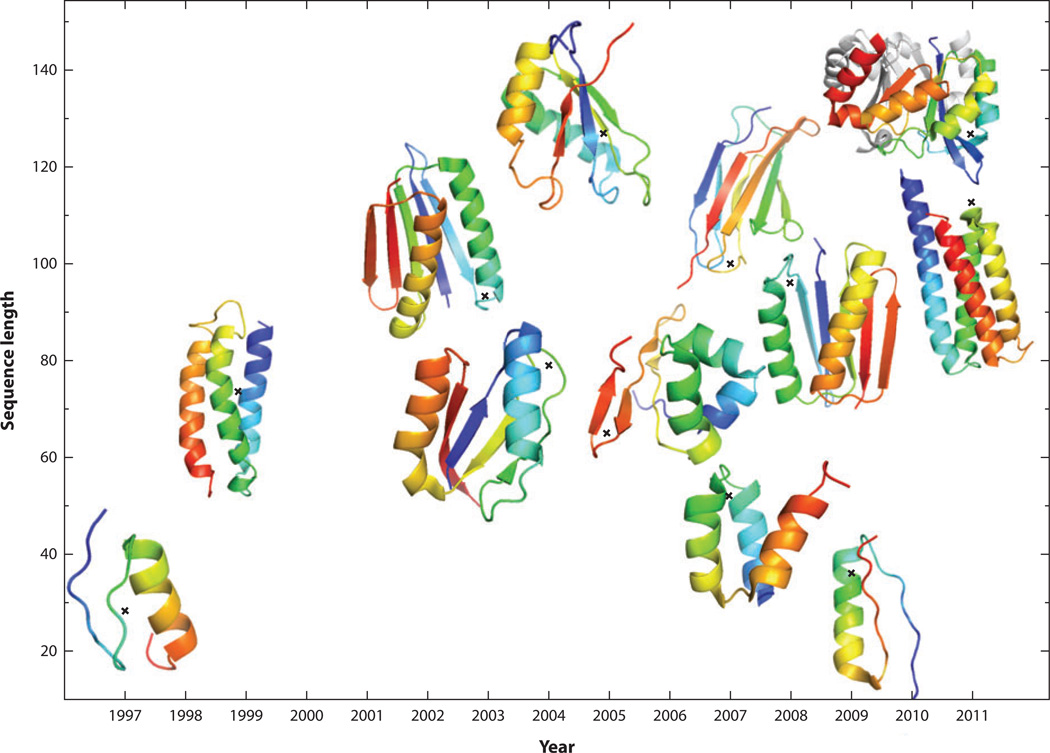
To retrieve all de novo designed and structurally validated proteins, we searched for the keywords “synthetic,” “de novo designed,” or “designed proteins” in the Protein Data Bank and excluded coiled coils, peptides, and those proteins that were not computationally designed (i.e., not designed by energy optimization). We also excluded those protein structures that have not been published in peer-reviewed publications. This leads to a small list of 12 proteins (see Table 1) whose structures were determined by NMR or X-ray diffraction over the span of 15 years. As shown in Figure 1, various structural folds have been successfully designed with increased complexities and sizes (defined as the number of amino acid residues); these range from all α, mixed α and β, and all β proteins. The largest computationally designed protein has 127 residues. Six of the 12 proteins listed were designed by RosettaDesign (21, 27, 36, 45, 58, 81), which utilized a mix of knowledge-based and physical-based energy terms with heavy emphasis on specific packing of hydrophobic and hydrophilic residues. The use of knowledge-based and/or physical-based energy functions for packing interactions is also crucial for other computational techniques (9, 17, 47, 65, 104, 108) to achieve structural specificity. However, over the past 15 years we have seen no significant change in the number of proteins that are de novo designed and structurally validated in a given year. It is either 0, 1, or 2 per year. This low number of designed proteins suggests lack of a broader utilization of computational design, lack of improvement in success rates, or both.
Table 1.
De novo, computationally designed proteins validated by NMR or X-ray structure determination
| Year | PDB ID | Length | Fold | Experiment | Computational method |
|---|---|---|---|---|---|
| 1997 | 1fsd | 28 | β−β−α motif | NMR | Pairwise residue-rotamer energy optimized by dead-end elimination (17) |
| 1999 | 2a3d | 73 | Three-helix bundle | NMR | Started from coiled coil, hydrophobic core repacked by a genetic algorithm (9) |
| 2003 | 1qys | 106 | Novel α+β | X-ray (1.2 Å) | Combining structure prediction with sequence design (RosettaDesign) (58) |
| 2004 | 1vjq | 79 | α/β | X-ray (2.1 Å) | RosettaDesign (21) |
| 2005 | 2cw1 | 65 | α+β | NMR | Optimizing a knowledge-based energy score by simulated annealing (47) |
| 2005 | 2a3j | 127 | α+β | NMR | RosettaDesign (27) |
| 2007 | 2p6j | 52 | Three-helix bundle | NMR | Fixed binary pattern, energy optimization by dead-end elimination, side chain conformations by Monte Carlo–simulated annealing (104) |
| 2007 | 3b83 | 100 | β-sandwich | X-ray (2.4 Å) | Energy function specifically optimized for β proteins (RosettaDesign) (45) |
| 2008 | 2jvf | 96 | α+β | NMR | Sequences generated from local tetrapeptide fragment library with some core residues fixed (108) |
| 2009 | 2ki0 | 36 | Novel β−α−β | NMR | A combination of knowledge-based secondary structure design with energy optimization (65) |
| 2011 | 3u3b | 113 | Four-helix bundle | X-ray (1.85 Å) | Allowed backbone flexibility for redesigning the entire hydrophobic core (RosettaDesign) (81) |
| 2011 | 3tdm | 126 | TIM-barrel α/β | X-ray (2 Å) | Imposing symmetry in RosettaDesign (36) |
Figure 1.
An increase in the sequence lengths of computationally designed and structurally validated proteins over the past 15 years.
ORIGIN OF LOW SUCCESS RATES IN PROTEIN DESIGN
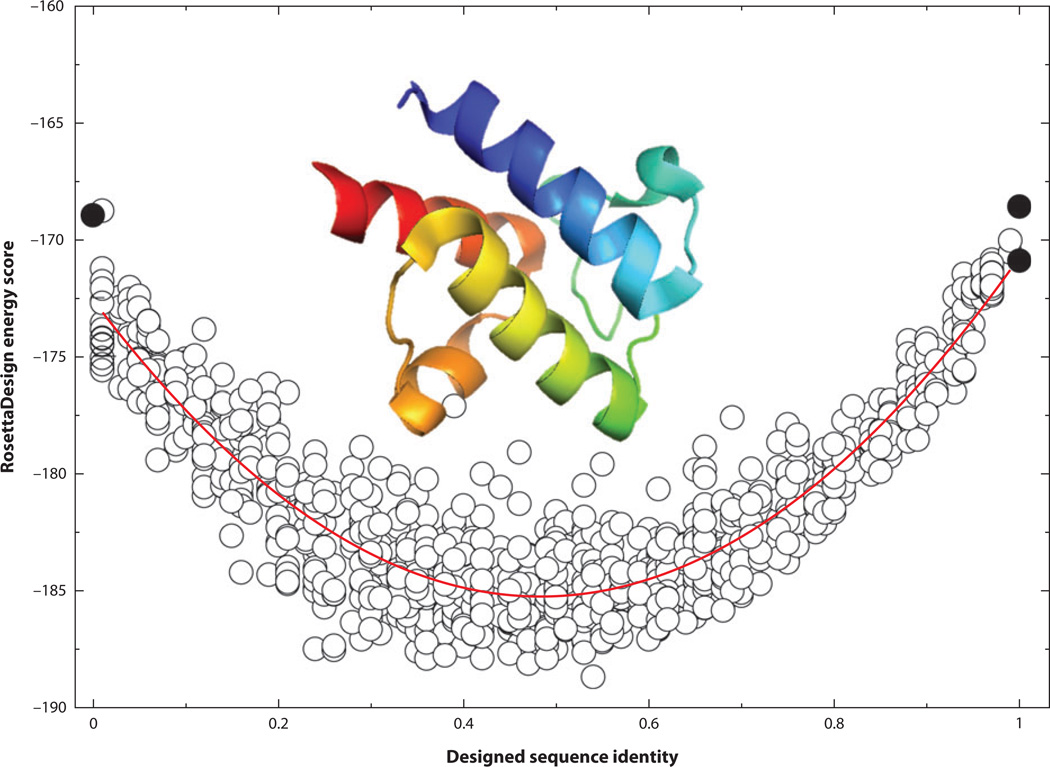
For a given protein length, an astronomically large number of possible sequences can be generated from different combinations of amino acid residues (20100 for a 100-residue protein). Only a tiny fraction of those sequences can be folded into specific structures by the water-mediated interactions among these residues. Thus, the observed low success rates in de novo design can be due to an inaccurate description of the interaction free energy function, a failure to locate the global minimum specified by the free energy function, or both. To assess which one is the likely cause, we examined 100 sequences designed by RosettaDesign 2.3 on the basis of different initial conditions. We (19) found that these sequences are highly homologous among each other, with an average sequence identity of 68% based on a database of 944 proteins. In other words, all designed sequences are converging around a single solution, suggesting that searching for a global minimum is not a major issue, at least for proteins designed with a fixed backbone. To confirm this, we added a harmonic restraint to the RosettaDesign energy function [E = −wseq(SeqID-SeqID0)2 with wseq = 10,000] so that we could sample sequences around a fixed sequence identity (SeqID0) to the wild-type sequence of the target structure. Figure 2 shows the RosettaDesign energy scores for 1,010 sequences designed for the structure of the acyl carrier protein from Thermus thermophilus HB8 (PDB ID: 1×3o) at different SeqID0 ranging from 0 to 1 (100%). Without the harmonic restraint, the average sequence identity to the wild-type sequence of the acyl carrier protein is around 50%. The energy score increases significantly when sequence identity moves toward either 0% or 100% sequence identity. This finding indicates that the wild-type sequence is not part of the solution. Because each RosettaDesign energy unit is 0.5–1 kcal mol−1 according to some estimates (23, 44, 106), the energy difference between the sequence at 100% sequence identity and the sequence at 50% sequence identity is about 15 RosettaDesign energy units or approximately 8–15 kcal mol−1. Although a wild-type sequence is not necessarily optimized for its structure, this energy difference is too large to be true because it is close to the typical stability free energy of proteins (−10 kcal mol−1) (90). The limitation of existing energy functions is further reflected from poorer performance in designing for NMR structures than for X-ray structures (57, 102). In other words, the quality of an energy function remains the main obstacle to successful computational design.
Figure 2.
The RosettaDesign energy score (RosettaDesign 2.3) as a function of sequence identity from the wild-type sequence of the acyl carrier protein from Thermus thermophilus HB8 (PDB ID: 1×3o). Different sequence identities were sampled by a harmonic restraint. The curved red line indicates the quadratic fit. Black circles at 100% sequence identity represent the energy value of the native structure with its wild-type sequence after side chain optimization from RosettaDesign (bottom), and the average energy value from 10 designed structures from RosettaDesign after fixing all residues to wild-type sequences without a harmonic restraint (top). The black circle at 0% sequence identity is the average energy value of 10 designed structures from RosettaDesign after excluding the type of wild-type amino acid residue at each sequence position without the harmonic restraint.
ENERGY FUNCTIONS FOR PROTEIN DESIGN
Energy functions for protein design are typically modified from the energy functions for protein folding or dynamics studies (for a discussion, see 6, 13, 72, 85, 88, 97, 109, 114). Because no major change in energy functions for protein design has occurred in the past decade, we do not provide a comprehensive summary of all existing energy functions employed in protein design. Instead, we describe in detail the energy functions of three programs [RosettaDesign (49, 57, 94), EGAD (87), and Liang-Grishin (66)], which are fairly representative of current state-of-the-art energy functions. RosettaDesign is dominated by knowledge-based energy functions derived from protein structures, with the exception of van der Waals and hydrogen bonding terms. EGAD attempts to build its energy function largely on a physical-based molecular mechanics force field. The Liang-Grishin scoring function, on the other hand, is an empirical mix of various geometry-based, knowledge-based, and physical-based terms. More importantly, these programs are available for our comparative studies.
RosettaDesign Energy Function
The RosettaDesign energy function (49, 57, 94) is made of 14 terms as shown in Equation 1 below:
| 1. |
where Eref and W are the optimized reference energy and weight factors for different energy terms, respectively. Eback is a backbone energy term for ϕ and ψ angles based on the Ramachandran diagram (75). is a statistical ω-angle potential. Erotamer is a backbone-dependent side chain rotamer energy term (28), which is a knowledge-based self-energy of an amino acid residue at a specific rotameric state derived from known protein structures. Eattr and Erepul are attractive and repulsive portions of the 12–6 Lennard-Jones potential, respectively. Erepul is finite and linearly dependent on distance for rij < 0.89σij (rij and σij are the distance between atoms i and j and the average van der Waals radius of atoms i and j, respectively). Intraresidue repulsive interactions are weighted separately. Esolv is the Lazaridis-Karplus implicit solvation energy (64). Ekele is a knowledge-based, electrostatic interaction based on the probability of two polar amino acid residues at a given distance (107). Ehbond is a geometry-based hydrogen bonding term that is weighted separately for local backbone-backbone (lb), nonlocal backbone-backbone (nlb), side chain–backbone (scb), and side chain–side chain (sc), respectively. EPro is a specific energy term for proline ring closure. There are also four additional terms for disulfide bonds. We do not list them here because RosettaDesign typically fixes Cys residues. All parameters and reference state values were optimized by native sequence recovery and amino acid compositions. Here we employ RosettaDesign 2.3 only because more recent versions do not make significant changes to its energy function.
EGAD Energy Function
An EGAD energy function (87) contains four terms:
| 2. |
where T is temperature. EOPLS-AA is the molecular mechanics energy function from the OPLS-AA (optimized potentials for liquid simulations–all atom) force field (51) that includes a van der Waals term, the Coulombic interaction, and torsion-angle terms, as well as truncated electrostatic energies between close atom pairs and a finite, linear repulsive term for the van der Waals interaction at rij < 0.82σij. The purpose of modification was to reduce hard-core overlap energies due to approximations introduced from fixed backbone and discrete side chain rotamers as in RosettaDesign. Esolv is the solvation free energy from the generalized Born model for electrostatic interactions and a solvent-accessible, surface-area-dependent term for hydrophobic interactions (86). Eref is a reference state energy estimated from the average of interaction energies for a given residue type in random sequences threaded onto protein structures. Sunfolded is side chain entropy (dependent on residue types only) in the unfolded state estimated from peptide simulations and rotamer statistics (15). In this equation, only two parameters for softening van der Waals repulsions were optimized for reproducing experimental mutation-induced change in protein stability. Pro, Gly, and Cys residues are fixed in the program.
Liang-Grishin Energy Function
The energy function for the Liang-Grishin method (66) is shown in Equation 3:
| 3. |
where Eref and W are the optimized reference energy and weight factors for different energy terms, respectively. Esurface and Evolume are the contacting surface area and overlapping volume between a rotamer and surrounding protein atoms (68), respectively. Ehbond is an empirical, geometry-based hydrogen bond energy function. Eelec represents CHARMM electrostatic interactions based on distance-dependent dielectric constants (8). Four desolvation energy terms are based on the buried hydrophobic surface area (), the hydrophilic surface area (), the fraction of buried surface area of non-hydrogen-bonded hydrophilic atoms ( ), and the solvent-exclusion volume of charged atoms Vexcl. Erotamer is an intrinsic rotamer energy term calculated on the basis of the expected rotamer frequency for a given amino acid residue type multiplied by the frequency of that amino acid type for given backbone torsion angles. The program also utilizes a specific disulfide bond term based on the number of disulfide bonds (Nssbond). All parameters and reference state values were optimized by native sequence recovery and amino acid compositions as in RosettaDesign.
Balancing Nonlocal and Local Interactions
All three energy functions, similar to other energy functions for protein design (6, 13, 72, 85, 88, 97, 109, 114), heavily emphasize nonlocal interactions between residues that are located close to each other in the three-dimensional space but far from each other in sequence positions. These nonlocal interactions (including van der Waals, electrostatic, hydrogen bonding, and solvation energies) were built for capturing tight and specific tertiary packing interactions. By comparison, local interactions between neighboring residues along a protein sequence are limited to a single-residue property such as secondary structure propensity as used in ORBIT (17), backbone torsion-angle terms (RosettaDesign and EGAD), and backbone torsion-angle-dependent rotamer energy (RosettaDesign and Liang-Grishin). On the other hand, secondary structures (or backbone torsion angles) are determined largely by a local sequence segment of 20 residues at about 80% accuracy for three-state secondary structures (32, 96) or 83% accuracy for both backbone φ and ψ torsion angles within 60° of their native values (31). Thus, going beyond single-residue properties may be required to account for the coupling between local backbone structures and sequences for protein design.
RosettaDesign-SR Energy Function
In order to examine the effect of local sequence-structure coupling, we modified the RosettaDesign energy function by adding three additional terms (19):
| 4. |
where Pprofile(i, Ii) is a structure-derived sequence profile (probability of an amino acid residue type, I, at a given sequence position, i). This sequence profile is generated by using target structural fragments to search for matching structural fragments stored in a fragments library. The sequences of the matching structural fragments are used to produce the probability of a given amino acid residue type at a given sequence position. The sequence profile for the whole target structure can be produced by a sliding window from the N terminus to the C terminus. This structure-derived sequence profile was successfully employed for protein structure prediction (121) and protein design (112). This profile term, however, leads to an increase in the number of repeats of same residue types, such as LLL and VVV, and a reduction in the complexity of designed sequences. Because protein sequences of low complexity are often associated with intrinsically disordered regions of a protein (95), such a low-complexity region is not desirable in designing structured proteins. Thus, to penalize a repetitive sequence segment, the second term in Equation 4 was introduced by calculating , the number of nearest and second-nearest neighboring residues (i − 2, i − 1, i + 1, i + 2) that repeat the residue type at the sequence position i. This second term is a simplified measure of the extent of sequence randomness by Shannon’s entropy (117). The third term in Equation 4 reflects the change to the reference state energy due to the introduction of new energy terms.
COMPUTATIONAL ASSESSMENT OF DESIGNED PROTEINS
How to make an accurate computational assessment of designed sequences remains an unsolved problem. We attempted to assess RosettaDesign, EGAD, Liang-Grishin, and RosettaDesign-SR (structure-derived sequence profile and repetitive penalty) on the basis of several criteria by employing a dataset of monomeric proteins to avoid possible complications due to interprotein interactions. The stably folded monomeric proteins are obtained by searching the Protein Data Bank for the following criteria: (a) X-ray-determined structures without DNA, RNA, hybrid, or other ligands; (b) proteins having only one chain (both biological assembly and asymmetric unit); (c) high resolution (≤3.0 Å), with the number of residues ≥70 and ≤400; and (d) proteins with no missing residues (except terminal regions) or abnormal amino acid types. A total of 616 proteins were obtained after removing redundant chains at 30% sequence identity. These proteins were then clustered according to their fractions of surface residues (fsr) in all amino acid residues of a protein, because proteins with more surface residues are more difficult to design owing to larger conformational freedom and more direct interactions with solvent molecules. We defined a residue as “on surface” if its solvent-accessible surface is greater than or equal to 20% of its reference value (99). We divided the target proteins according to the ranges of their fsr values {[0.4, 0.45), [0.45, 0.5), [0.5, 0.55), [0.55, 0.6), [0.6, 0.65), [0.65, 0.7), [0.7, 0.75), and [0.75–0.85)}. We started from 0.4 because few proteins have fsr values less than 0.4. For the same reason, the last bin was combined from two bins, [0.75–0.8) and [0.8–0.85). Because designing proteins with EGAD and Liang-Grishin programs is computationally intensive, we only designed 15 of the smallest proteins per bin, although the last bin had only 7 proteins, all from the dataset of 616 proteins. A total of 112 proteins were designed by four programs. (The list of 112 proteins is provided at http://sparks.informatics.iupui.edu.) We employed all default settings in those programs for fixed backbone design to increase computational efficiency, and removed all side chains from these structures prior to computational design.
Sequence Assessment: Native Sequence Recovery
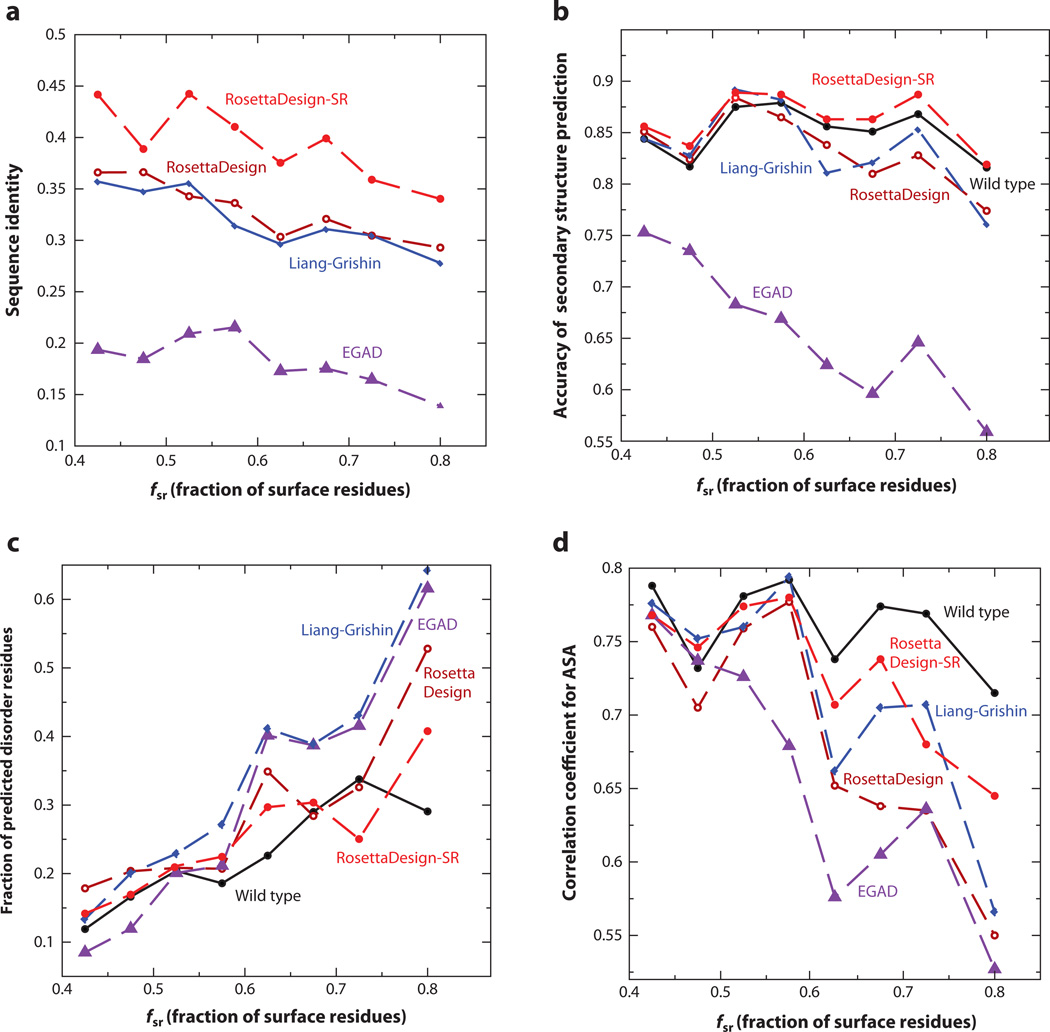
One commonly employed approach for assessing designed sequences is to calculate the sequence identity to the wild-type (or native) sequence or the recovery rate of the native sequence for a given target structure. The reported sequence identities range from 30% to 37% (45, 57, 66, 73, 87). These results were often based on a small number of proteins. Moreover, some methods fixed certain types of amino acid residues such as Gly, Cys, or Pro. Figure 3a compares the average sequence identity of designed sequences to their respective wild-type sequences at different fractions of surface residues without fixing any residue types. RosettaDesign-SR gives the highest sequence identities, ranging from 36% to 44%, which are 4–8% better than the next best shared by RosettaDesign and Liang-Grishin that yield similar sequence identities. The lowest sequence identity was given by EGAD in all methods examined, likely because EGAD was not optimized for the native sequence recovery.
Figure 3.
(a) The average sequence identity of sequences designed by RosettaDesign-SR, RosettaDesign, Liang-Grishin, and EGAD is compared to their respective wild-type sequences as a function of the fraction of surface residues. (b) The average accuracy of predicted secondary structures from the sequences designed by four computational methods is compared with the results for wild-type sequences. SPINE-X was employed for sequence-based secondary structure prediction. (c) The average fractions of predicted disordered residues are compared. SPINE-D was employed for predicting intrinsic disorder for designed and wild-type sequences. (d) The average correlation coefficients between predicted and actual solvent-accessible surface areas (ASA) from the target structure are compared. Real-SPINE 3 was employed for solvent accessibility prediction from designed and wild-type sequences.
Local Assessment: Secondary Structure Recovery
The effect of lacking the local coupling term between sequence and backbone structure can be examined by comparing the accuracy of predicted secondary structures for designed sequences or the ability of recovering native secondary structures. We employed SPINE-X for secondary structure prediction, which achieves 81–82% accuracy for large benchmark tests (32). Figure 3b shows that the average accuracy of predicted secondary structures for sequences designed by RosettaDesign-SR is consistently higher than the accuracy of structures predicted from wild-type sequences. This reflects the usefulness of utilizing the local-structure-derived sequence profile in RosettaDesign-SR. The sequences designed by the RosettaDesign and Liang-Grishin programs yielded more accurate secondary structures than did wild-type sequences at low fractions of surface residues but not at high fractions of surface residues. This suggests that local sequence-structure coupling is more effective for capturing correct secondary structures in surface regions. EGAD has the lowest recovery of native secondary structures, consistent with its low sequence identity to wild-type sequences.
Local Assessment: Predicted Intrinsic Disorder
The possibility of low complexity in designed sequences leads us to examine predicted intrinsically disordered residues in designed sequences. We employ SPINE-D (120) for this task because it was one of the top disorder predictors in critical assessments of structure prediction techniques in 2010 (CASP 9) (79). Figure 3c compares average fractions of disordered residues given by wild-type sequences with those from designed sequences at different fractions of surface residues. Except for one bin where a few wild-type sequences have regions with predicted disorder probabilities at about 0.5, the fractions of disordered residues in wild-type sequences are usually lower than those in designed sequences. This suggests the usefulness of SPINE-D for detecting potentially unstable regions of designed sequences. Liang-Grishin and EGAD programs yielded sequences with higher fractions of predicted disordered residues than did wild-type sequences, whereas the sequences generated from RosettaDesign-SR and RosettaDesign programs and wild-type sequences have a similar amount of disorder in most bins.
Surface Assessment: Solvent Accessibility Recovery
Another way to examine designed sequences is to test the conservation of solvent-accessible surface area (ASA) of designed sequences relative to that of native structures of wild-type sequences. We calculated the correlation coefficient between the ASA predicted by Real-SPINE 3 (30) and actual ASA based on the corresponding wild-type sequence on the target structure. Figure 3d shows that at low fractions of surface residues, all sequences yield similar correlation coefficients for ASA (~0.75). The difference between different methods increases for proteins with higher fractions of surface residues. Sequences designed by RosettaDesign-SR and Liang-Grishin programs produced ASA closer to that of wild-type sequences than did RosettaDesign and EGAD programs.
Surface Assessment: Hydrophobic Patch
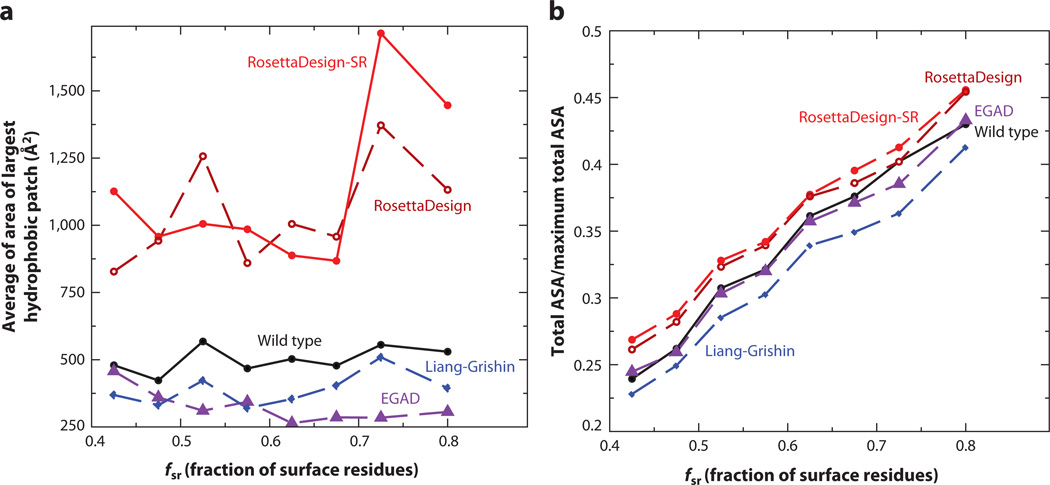
Aggregation is one common problem for designed proteins (103). Rate of aggregation is associated with exposed hydrophobic surface areas (11). Figure 4a compares the average largest hydrophobic patch area given by different methods. The hydrophobic surface patch area on a target structure with a designed sequence was generated by the program QUILT (70). RosettaDesign and RosettaDesign-SR programs produced significantly higher hydrophobic patch areas (2–3 times higher) than wild-type proteins do. Remarkably, the sequences designed by the Liang-Grishin program have smaller hydrophobic patch areas than the wild-type sequences. This finding highlights the emphasis of the Liang-Grishin energy function on surface-exposed residues with four separate solvation terms. EGAD-designed proteins also produced smaller hydrophobic patches than wild-type proteins. One should note, however, that designed sequences with large hydrophobic patches may be filtered by manual selection of sequences for experimental validations.
Figure 4.
(a) A comparison of the average largest hydrophobic patch area given by RosettaDesign-SR, RosettaDesign, Liang-Grishin, and EGAD with that given by wild-type proteins. (b) A comparison of the total solvent-accessible surface area (ASA) for all residues in a protein normalized by their maximum possible total solvent-accessible surface area for the four programs and wild type.
Packing Assessment: Total Accessible Surface Area
Packing interaction is the dominant stabilization factor for specific tertiary structures. We utilized the target structure with designed sequences to calculate total solvent accessible surface areas for all residues in a protein normalized by their maximum total (reference) solvent-accessible area. Figure 4b shows that RosettaDesign and RosettaDesign-SR programs yielded higher values (about 8%) of total ASA than wild-type sequences did, whereas the Liang-Grishin program gave significantly lower values of total ASA. The EGAD program, on the other hand, yielded ASA values essentially equal to those of wild-type sequences. This suggests that protein cores designed by RosettaDesign and RosettaDesign-SR do not pack as tightly as EGAD and native proteins. The Liang-Grishin program seems to pack protein cores more tightly than native proteins.
Global Structure Assessment
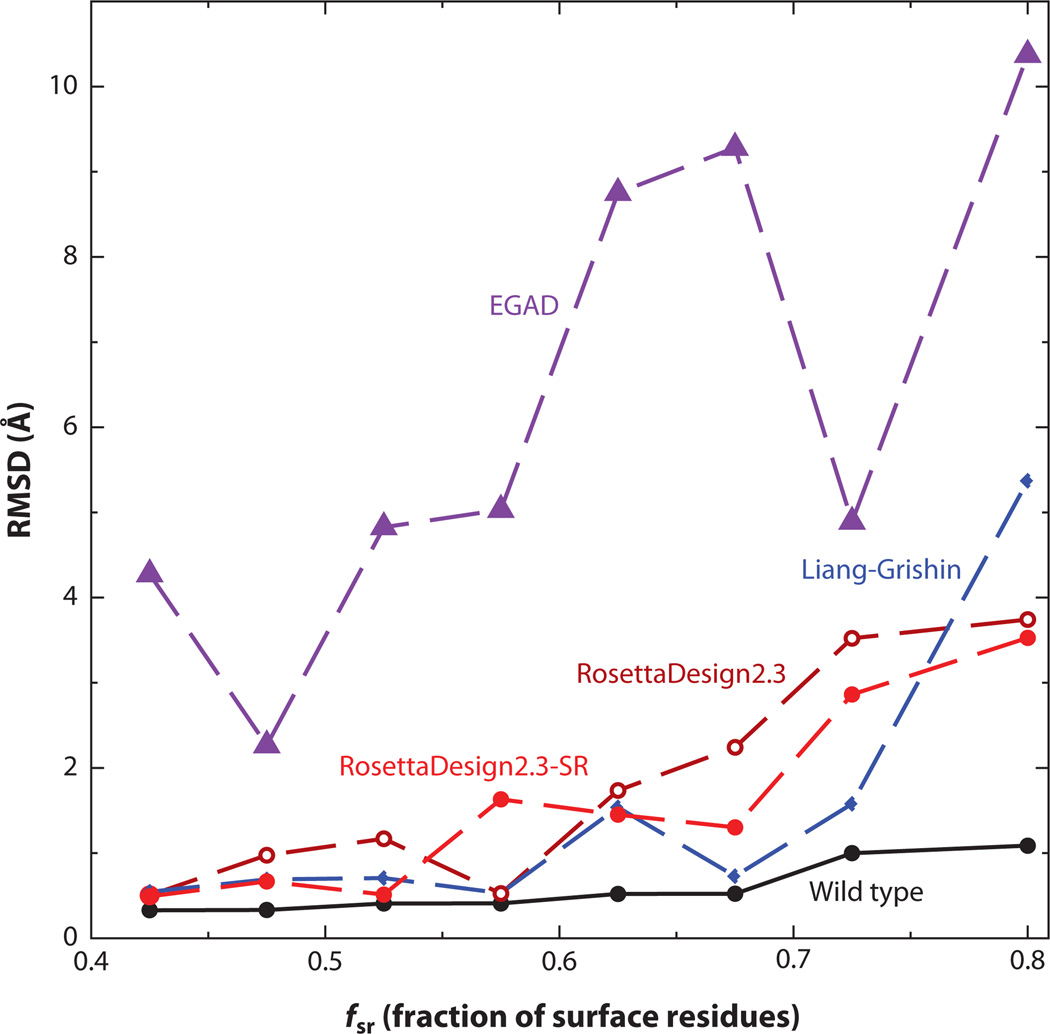
Designed sequences can also be assessed globally. One method to examine the stabilities of designed proteins is to perform molecular dynamics simulations. A stably folded protein is expected to maintain its structure after a long molecular dynamics simulation. For example, Tsai et al. (112) designed two proteins (proteinGB1 domain and ubiquitin) by combinatorial assembly of fragments in the Protein Data Bank. Stabilities of designed proteins were evaluated by molecular dynamics simulations. Designed proteins for protein GB1 domain and ubiquitin have higher root-mean-squared distances (RMSD) from the target structure than wild-type proteins but lower RMSD than nonprotein controls (inverted hydrophobic/hydrophilic residue patterns). Liang et al. (69) designed protein-protein interaction interfaces by grafting binding epitopes onto small proteins. Molecular dynamics simulations revealed that some designed interfaces are not stable (disassociating) during the course of long molecular dynamics simulations whereas interfaces and natively binding proteins remain stable. Another way to assess designed proteins globally is to predict structures of designed sequences. For example, Bazzoli et al. (4) assessed designed sequences using the fragment/template-based structure prediction technique I-TASSER. They found that the majority of top designed sequences have folded into the structures within 2 Å RMSD from the target structure, even though different energy-scoring functions were used in design and folding assembly. Here, we (118) employed the template-based structure prediction tool SPARKS-X to predict structures of designed sequences where the target structures are contained in the template library. The predicted structures were then compared to their respective target structures by RMSD. Figure 5 shows that the performances of Liang-Grishin, RosettaDesign, and RosettaDesign-SR programs are similar. EGAD performed the worst largely because its low native sequence recovery makes recognizing correct template structures difficult. Note that even wild-type sequences have small RMSD values because SPARKS-X rebuilt and refined predicted structures using the program MODELLER (98).
Figure 5.
The average root-mean-squared distance (RMSD) between the target structure and the structure predicted by the template-based structure prediction method SPARKS-X, based on designed sequences at different fractions of surface residues.
Summary
On the basis of the results from Figures 3, 4, and 5, it is clear that introducing local sequence structure coupling and sequence complexity terms into RosettaDesign (RosettaDesign-SR) leads to the intended effect of increasing sequence identity to wild-type sequence (Figure 3a) and improving the consistency between predicted secondary structure and actual secondary structure (Figure 3b), and between predicted ASA and actual ASA (Figure 3d). However, the average largest hydrophobic patch area given by RosettaDesign-SR, as by RosettaDesign, is too large, compared with that given by wild-type sequences. This result points out an area for future improvement by introducing explicit (18, 49, 110) or implicit (116) scoring methods for hydrophobic patches. Although reference energies, in principle, can control the amount of the hydrophobic surface area exposed by controlling the ratio of hydrophobic to hydrophilic residues, such reference states do not seem adequate in RosettaDesign or RosettaDesign-SR. Another interesting result is that Liang-Grishin and EGAD programs performed the best in terms of sizes of the largest hydrophobic patch. However, too few hydrophobic residues on the surface may reduce the overall stability of proteins because hydrophobic interactions are the major driving force of protein stability (26). Even surface hydrophobic residues improve protein stability (89, 101). Thus, weighting various energetic terms differently leads to different outcomes. Determining how to balance these different interactions is the key to successful protein design.
COMMUNITY-WIDE SCORING FUNCTION ASSESSMENT
Recently, a large number of designed proteins targeting the conserved stem region of influenza hemagglutinin (34) offered an unprecedented opportunity to examine the ability of energy-scoring functions to separate binders from nonbinders by a blind-prediction, community-wide experiment (35). Twenty-eight groups, including ours, armed with different energy functions participated in this experiment. These energy functions range from physical-based molecular mechanics force fields, knowledge-based energy functions, empirical combinations of various knowledge-based and physical-based terms, to scoring functions trained by machine learning techniques. The highest area under the receiver operating characteristic curve for two-state binding/nonbinding prediction is 0.86 by three scoring functions. Two scoring functions (Group 2 by J.C. Mitchell & O.N.A. Demerdash and Group 6 by I.H. Moal, X. Li & P.A. Bates) are specifically trained for binding/nonbinding classification by employing support vector machines (SVM) with many knowledge-based and physical-based features. The third scoring function (Group 7 by M. Zacharias) is a coarse-grained force field with energy parameters optimized for scoring near-native docking decoys (33). Yet, these best energy-scoring functions failed to adequately separate native from designed interfaces and to identify an experimentally validated designed binder (35). Thus, it is difficult to assess what really worked for these best energy-scoring functions except that specific training is needed for balancing the terms in the scoring functions.
CURRENT CHALLENGES AND FUTURE PROSPECTS
The above assessment of designed sequences highlights the importance of balancing different types of interactions. Folded and functional proteins result from the interplay of backbone and side chain interactions and delicate balance among van der Waals interactions, electrostatic interactions, and solvation effects. Nature has mastered the art of balance via trial and error over the course of billions of years. Furthermore, it employs quantum effects to enhance its magic. Various knowledge-based, physical-based, and empirical energy functions have been proposed over the years (6, 13, 72, 85, 88, 97, 109, 114, 123), including a recent solvent-exposure-dependent potential (25) and structure-derived sequence profile and sequence complexity (19). We believe that the next practical step for significantly improving protein design is not to search for new terms but to select the correct terms whose weights are optimized with appropriate objective functions. The usefulness of rebalancing energy terms is suggested from the success of employing SVM-trained scoring functions to separate binding from nonbinding designed interfaces (35) and of balancing local and nonlocal interactions to achieve higher recovery of native sequence, secondary structure, and solvent accessibility (19). Balancing stability and solubility (18, 49, 110) is another key aspect for producing functional and foldable globular proteins.
Our optimism for individual energy terms is built on the discovery that in some cases knowledge-based energy functions are directly comparable to quantum calculations. Examples include the agreement between a statistical hydrogen-bonding potential and quantum mechanical calculations (80) and the strong positive correlation between statistical descriptions of cation-π and amino-π interactions and quantum calculations at the Hartree-Fock and the second-order Möller-Plesset perturbation theory levels (38). In addition, recently developed, orientation-dependent (10, 67, 76, 119) and multibody (39) energy functions have yet to be tested for protein design. For example, the dipolar DFIRE (Distance-scaled, Finite, Ideal-gas REference) energy function (119) based on a DFIRE state (122) accounts for the orientation dependence of the interactions not only between hydrogen-bonded polar atoms but also between other polar atoms and between polar atoms and nonpolar atoms. The last interaction is known to play important role in secondary structure formation (24, 77, 83).
There is another balance that needs attention: the balance of speed and accuracy. Fixed backbone structures were employed for all tests performed here in order to reduce computing time. Fixing backbone structures may have made protein structures less favorable to native sequences as a result of employing less accurate energy functions for compensating the effects of rigid backbone and discretization of side chain conformations. Allowing flexibility improved sequence identity between designed and wild-type sequences (100) and in successful redesign of the hydrophobic core (81). Discretization of side chain rotamers is another issue that may adversely affect the performance of an energy function. Gainza et al. (37) showed that employing continuous rotamers leads to an impressive 10% improvement in sequence identity by redesigning 12–15 selected core residues. That is, not all problems in protein design are caused by defects in energy functions. Unfortunately, efficient sampling of the conformational space of flexible proteins has not been resolved, although progress has been made (78).
The main obstacle to searching for the right balance of correct terms in energy functions is the lack of a large number of negative experiments for understanding where designs have failed and for training the delicate balance of various energetic terms. This lack is caused by two factors. First, most publications reported only successfully designed sequences. Second, few laboratories can afford a large number of experiments to measure the success rate of protein design. The large number of designed proteins targeting influenza hemagglutinin (34) is the first sizeable dataset of negative examples for protein-protein interactions. Experiments such as this in de novo protein design are needed to further understand deficiencies in existing energy-scoring functions and to achieve the optimal balance between selected energetic terms. This balance will happen when inexpensive high-throughput techniques for measuring the success rate of protein design become available.
SUMMARY POINTS.
Low success rate is due largely to poor energy functions employed for protein design.
The quality of an energy function can be significantly improved by locating the correct interaction terms and optimizing their weights.
The correct balance of interaction terms may be found by incorporating experimental negative data into training.
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM085003. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Helpful discussions with Ken Dill, Amy Keating, and Shide Liang are gratefully acknowledged.
Glossary
- De novo protein design
computationally designed proteins that can fold into a target structure with a desired function
- Intrinsic disorder
proteins or regions in a protein that do not have a unique three-dimensional structure as a monomer at physiological conditions
- Knowledge-based energy function
an energy function derived from statistical or statistical mechanical analysis of known protein structures
- Physical-based energy function
an energy function derived by the laws of physics that is composed of many approximate terms
- Energy function
the scoring function that is minimized during iterative protein design
- Local interaction
the interaction between amino acid residues that are sequence neighbors
- Nonlocal interaction
the interaction between amino acid residues that are located close to each other in three-dimensional space but far from each other in their sequence positions
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Ambroggio XI, Kuhlman B. Computational design of a single amino acid sequence that can switch between two distinct protein folds. J. Am. Chem. Soc. 2006;128:1154–1161. doi: 10.1021/ja054718w. [DOI] [PubMed] [Google Scholar]
- 2.Ambroggio XI, Kuhlman B. Design of protein conformational switches. Curr. Opin. Struct. Biol. 2006;16:525–530. doi: 10.1016/j.sbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ, et al. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazzoli A, Tettamanzi AGB, Zhang Y. Computational protein design and large-scale assessment by I-TASSER structure assembly simulations. J. Mol. Biol. 2011;407:764–776. doi: 10.1016/j.jmb.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender GM, Lehmann A, Zou H, Cheng H, Fry HC, et al. De novo design of a single-chain diphenylporphyrin metalloprotein. J. Am. Chem. Soc. 2007;129:10732–10740. doi: 10.1021/ja071199j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boas FE, Harbury PB. Potential energy functions for protein design. Curr. Opin. Struct. Biol. 2007;17:199–204. doi: 10.1016/j.sbi.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Bolon DN, Voigt CA, Mayo SL. De novo design of biocatalysts. Curr. Opin. Chem. Biol. 2002;6:125–129. doi: 10.1016/s1367-5931(02)00303-4. [DOI] [PubMed] [Google Scholar]
- 8.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 9.Bryson JW, Desjarlais JR, Handel TM, DeGrado WF. From coiled coils to small globular proteins: design of a native-like three-helix bundle. Protein Sci. 1998;7:1404–1414. doi: 10.1002/pro.5560070617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchete NV, Straub JE, Thirumalai D. Development of novel statistical potentials for protein fold recognition. Curr. Opin. Struct. Biol. 2004;14:225–232. doi: 10.1016/j.sbi.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 12.Clark LA, Boriack-Sjodin PA, Eldredge J, Fitch C, Friedman B, et al. Affinity enhancement of an in vivo matured therapeutic antibody using structure-based computational design. Protein Sci. 2006;15:949–960. doi: 10.1110/ps.052030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cootes AP, Curmi PMG, Torda AE. Automated protein design and sequence optimisation: scoring functions and the search problem. Curr. Protein Pept. Sci. 2000;1:255–271. doi: 10.2174/1389203003381351. [DOI] [PubMed] [Google Scholar]
- 14.Cossio P, Trovato A, Pietrucci F, Seno F, Maritan A, Laio A. Exploring the universe of protein structures beyond the Protein Data Bank. PLoS Comput. Biol. 2010;6:E1000957. doi: 10.1371/journal.pcbi.1000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Creamer TP. Side-chain conformational entropy in protein unfolded states. Proteins. 2000;40:443–450. doi: 10.1002/1097-0134(20000815)40:3<443::aid-prot100>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 16.Cunningham BA, Hemperly JJ, Hopp TP, Edelman GM. Favin versus concanavalin A: circularly permuted amino acid sequences. Proc. Natl. Acad. Sci. USA. 1979;76:3218–3222. doi: 10.1073/pnas.76.7.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dahiyat BI, Mayo SL. De novo protein design: fully automated sequence selection. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. Designed structure validated by NMR structure determination for the first time.
- 18.Dahiyat BI, Mayo SL. Probing the role of packing specificity in protein design. Proc. Natl. Acad. Sci. USA. 1997;94:10172–10177. doi: 10.1073/pnas.94.19.10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dai L, Yang Y, Kim HR, Zhou Y. Improving computational protein design by using structure-derived sequence profile. Proteins. 2010;78:2338–2348. doi: 10.1002/prot.22746. Discussed the importance of local interactions for protein design.
- 20. Dai L, Zhou Y. Characterizing the existing and potential structural space of proteins by large-scale multiple loop permutations. J. Mol. Biol. 2011;408:585–595. doi: 10.1016/j.jmb.2011.02.056. Designed novel structures by a computational multiloop permutation technique.
- 21.Dantas G, Corrent C, Reichow SL, Havranek JJ, Eletr ZM, et al. High-resolution structural and thermodynamic analysis of extreme stabilization of human procarboxypeptidase by computational protein design. J. Mol. Biol. 2007;366:1209–1221. doi: 10.1016/j.jmb.2006.11.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dantas G, Kuhlman B, Callender D, Wong M, Baker D. A large scale test of computational protein design: folding and stability of nine completely redesigned globular proteins. J. Mol. Biol. 2003;332:449–460. doi: 10.1016/s0022-2836(03)00888-x. [DOI] [PubMed] [Google Scholar]
- 23.Das R. Four small puzzles that Rosetta doesn’t solve. PLoS ONE. 2011;6:e20044. doi: 10.1371/journal.pone.0020044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deane CM, Allen FH, Taylor R, Blundell TL. Carbonyl-carbonyl interactions stabilize the partially allowed Ramachandran conformations of asparagine and aspartic acid. Protein Eng. 1999;12:1025–1028. doi: 10.1093/protein/12.12.1025. [DOI] [PubMed] [Google Scholar]
- 25.DeLuca S, Dorr B, Meiler J. Design of native-like proteins through an exposure-dependent environment potential. Biochemistry. 2011;50:8521–8528. doi: 10.1021/bi200664b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dill KA, Stigter D. Modeling protein stability as heteropolymer collapse. Adv. Protein Chem. 1995;46:59–104. doi: 10.1016/s0065-3233(08)60332-0. [DOI] [PubMed] [Google Scholar]
- 27.Dobson N, Dantas G, Baker D, Varani G. High-resolution structural validation of the computational redesign of human U1A protein. Structure. 2006;14:847–856. doi: 10.1016/j.str.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 28.Dunbrack RL, Jr, Karplus M. Conformational analysis of the backbone-dependent rotamer preferences of protein sidechains. Nat. Struct. Biol. 1994;1:334–340. doi: 10.1038/nsb0594-334. [DOI] [PubMed] [Google Scholar]
- 29.Dwyer MA, Looger LL, Hellinga HW. Computational design of a biologically active enzyme. Science. 2004;304:1967–1971. doi: 10.1126/science.1098432. [DOI] [PubMed] [Google Scholar]
- 30.Faraggi E, Xue B, Zhou Y. Improving the prediction accuracy of residue solvent accessibility and real-value backbone torsion angles of proteins by guided-learning through a two-layer neural network. Proteins. 2009;74:847–856. doi: 10.1002/prot.22193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faraggi E, Yang YD, Zhang SS, Zhou Y. Predicting continuous local structure and the effect of its substitution for secondary structure in fragment-free protein structure prediction. Structure. 2009;17:1515–1527. doi: 10.1016/j.str.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faraggi E, Zhang T, Yang Y, Kurgan L, Zhou Y. SPINE X: improving protein secondary structure prediction by multi-step learning coupled with prediction of solvent accessible surface area and backbone torsion angles. J. Comput. Chem. 2011;33:259–263. doi: 10.1002/jcc.21968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiorucci S, Zacharias M. Binding site prediction and improved scoring during flexible protein-protein docking with ATTRACT. Proteins. 2010;78:3131–3139. doi: 10.1002/prot.22808. [DOI] [PubMed] [Google Scholar]
- 34.Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fleishman SJ, Whitehead TA, Strauch EM, Corn JE, Qin SB, et al. Community-wide assessment of protein-interface modeling suggests improvements to design methodology. J. Mol. Biol. 2011;414:289–302. doi: 10.1016/j.jmb.2011.09.031. Conducted a large-scale blind test of scoring functions for protein-interface design.
- 36.Fortenberry C, Bowman EA, Proffitt W, Dorr B, Combs S, et al. Exploring symmetry as an avenue to the computational design of large protein domains. J. Am. Chem. Soc. 2011;133:18026–18029. doi: 10.1021/ja2051217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gainza P, Roberts KE, Donald BR. Protein design using continuous rotamers. PLoS Comput. Biol. 2012;8:E1002335. doi: 10.1371/journal.pcbi.1002335. Discussed the importance of continuous space for side chains.
- 38.Gilis D, Biot C, Buisine E, Dehouck Y, Rooman M. Development of novel statistical potentials describing cation-πinteractions in proteins and comparison with semiempirical and quantum chemistry approaches. J. Chem. Inform. Model. 2006;46:884–893. doi: 10.1021/ci050395b. [DOI] [PubMed] [Google Scholar]
- 39.Gniewek P, Leelananda SP, Kolinski A, Jernigan RL, Kloczkowski A. Multibody coarse-grained potentials for native structure recognition and quality assessment of protein models. Proteins. 2011;79:1923–1929. doi: 10.1002/prot.23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS. High-resolution protein design with backbone freedom. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 42.Hennecke J, Sebbel P, Glockshuber R. Random circular permutation of DsbA reveals segments that are essential for protein folding and stability. J. Mol. Biol. 1999;286:1197–1215. doi: 10.1006/jmbi.1998.2531. [DOI] [PubMed] [Google Scholar]
- 43.Hill CP, Anderson DH, Wesson L, DeGrado WF, Eisenberg D. Crystal structure of alpha 1: implications for protein design. Science. 1990;249:543–546. doi: 10.1126/science.2382133. [DOI] [PubMed] [Google Scholar]
- 44.Hu XZ, Wang HC, Ke HM, Kuhlman B. High-resolution design of a protein loop. Proc. Natl. Acad. Sci. USA. 2007;104:17668–17673. doi: 10.1073/pnas.0707977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu XZ, Wang HC, Ke HM, Kuhlman B. Computer-based redesign of a β sandwich protein suggests that extensive negative design is not required for de novo β sheet design. Structure. 2008;16:1799–1805. doi: 10.1016/j.str.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang JK, Lai YL, Yen SC. Comprehensive analysis of knotted proteins. In: Zhao Z, editor. Sequence and Genome Analysis: Methods and Applications. Queensland, Aust: iConcept Press; 2010. pp. 22–39. [Google Scholar]
- 47.Isogai Y, Ito Y, Ikeya T, Shiro Y, Ota M. Design of λ Cro fold: solution structure of a monomeric variant of the de novo protein. J. Mol. Biol. 2005;354:801–814. doi: 10.1016/j.jmb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 48.Iwakura M, Nakamura T, Yamane C, Maki K. Systematic circular permutation of an entire protein reveals essential folding elements. Nat. Struct. Biol. 2000;7:580–585. doi: 10.1038/76811. [DOI] [PubMed] [Google Scholar]
- 49. Jacak R, Leaver-Fay A, Kuhlman B. Computational protein design with explicit consideration of surface hydrophobic patches. Proteins. 2012;80:825–838. doi: 10.1002/prot.23241. Introduced an explicit hydrophobic patch term.
- 50.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- 52.Kamtekar S, Schiffer JM, Xiong H, Babik JM, Hecht MH. Protein design by binary patterning of polar and nonpolar amino acids. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- 53.King NP, Jacobitz AW, Sawaya MR, Goldschmidt L, Yeates TO. Structure and folding of a designed knotted protein. Proc. Natl. Acad. Sci. USA. 2010;107:20732–20737. doi: 10.1073/pnas.1007602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koder RL, Anderson JL, Solomon LA, Reddy KS, Moser CC, Dutton PL. Design and engineering of an O2 transport protein. Nature. 2009;458:305–309. doi: 10.1038/nature07841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kolesov G, Virnau P, Kardar M, Mirny LA. Protein knot server: detection of knots in protein structures. Nucleic Acids Res. 2007;35:W425–W428. doi: 10.1093/nar/gkm312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kortemme T, Ramirez-Alvarado M, Serrano L. Design of a 20-amino acid, three-stranded beta-sheet protein. Science. 1998;281:253–256. doi: 10.1126/science.281.5374.253. [DOI] [PubMed] [Google Scholar]
- 57.Kuhlman B, Baker D. Native protein sequences are close to optimal for their structures. Proc. Natl. Acad. Sci. USA. 2000;97:13383–13388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. Designed a novel protein fold.
- 59.Kuhlman B, O’Neill JW, Kim DE, Zhang KY, Baker D. Conversion of monomeric protein L to an obligate dimer by computational protein design. Proc. Natl. Acad. Sci. USA. 2001;98:10687–10691. doi: 10.1073/pnas.181354398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuhlman B, O’Neill JW, Kim DE, Zhang KY, Baker D. Accurate computer-based design of a new backbone conformation in the second turn of protein L. J. Mol. Biol. 2002;315:471–477. doi: 10.1006/jmbi.2001.5229. [DOI] [PubMed] [Google Scholar]
- 61.Lai YL, Yen SC, Yu SH, Hwang JK. pKNOT: the protein KNOT web server. Nucleic Acids Res. 2007;35:W420–W424. doi: 10.1093/nar/gkm304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lassila JK, Keeffe JR, Oelschlaeger P, Mayo SL. Computationally designed variants of Escherichia coli chorismate mutase show altered catalytic activity. Protein Eng. Des. Sel. 2005;18:161–163. doi: 10.1093/protein/gzi015. [DOI] [PubMed] [Google Scholar]
- 63.Lazar GA, Dang W, Karki S, Vafa O, Peng JS, et al. Engineered antibody Fc variants with enhanced effector function. Proc. Natl. Acad. Sci. USA. 2006;103:4005–4010. doi: 10.1073/pnas.0508123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 65.Liang H, Chen H, Fan K, Wei P, Guo X, et al. De novo design of a βαβ motif. Angew. Chem. 2009;48:3301–3303. doi: 10.1002/anie.200805476. [DOI] [PubMed] [Google Scholar]
- 66.Liang S, Grishin NV. Effective scoring function for protein sequence design. Proteins. 2004;54:271–281. doi: 10.1002/prot.10560. [DOI] [PubMed] [Google Scholar]
- 67.Liang S, Zhou Y, Grishin N, Standley DM. Protein side chain modeling with orientation-dependent atomic force fields derived by series expansions. J. Comput. Chem. 2011;32:1680–1686. doi: 10.1002/jcc.21747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang SD, Grishin NV. Side-chain modeling with an optimized scoring function. Protein Sci. 2002;11:322–331. doi: 10.1110/ps.24902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang SD, Li LW, Hsu WL, Pilcher MN, Uversky V, et al. Exploring the molecular design of protein interaction sites with molecular dynamics simulations and free energy calculations. Biochemistry. 2009;48:399–414. doi: 10.1021/bi8017043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lijnzaad P, Berendsen HJ, Argos P. A method for detecting hydrophobic patches on protein surfaces. Proteins. 1996;26:192–203. doi: 10.1002/(SICI)1097-0134(199610)26:2<192::AID-PROT9>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 71.Lindqvist Y, Schneider G. Circular permutations of natural protein sequences: structural evidence. Curr. Opin. Struct. Biol. 1997;7:422–427. doi: 10.1016/s0959-440x(97)80061-9. [DOI] [PubMed] [Google Scholar]
- 72.Lippow SM, Tidor B. Progress in computational protein design. Curr. Opin. Biotechnol. 2007;18:305–311. doi: 10.1016/j.copbio.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Y, Kuhlman B. RosettaDesign server for protein design. Nucleic Acids Res. 2006;34:W235–W238. doi: 10.1093/nar/gkl163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Looger LL, Dwyer MA, Smith JJ, Hellinga HW. Computational design of receptor and sensor proteins with novel functions. Nature. 2003;423:185–190. doi: 10.1038/nature01556. [DOI] [PubMed] [Google Scholar]
- 75.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, et al. Structure validation by Cαgeometry: ϕ, ψand Cβdeviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 76.Ma J. Explicit orientation dependence in empirical potentials and its significance to side-chain modeling. Acc. Chem. Res. 2009;42:1087–1096. doi: 10.1021/ar900009e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maccallum PH, Poet R, JamesMilner-White E. Coulombic interactions between partially charged main-chain atoms not hydrogen-bonded to each other influence the conformations of α-helices and antiparallelβ-sheet.Anewmethod for analysing the forces between hydrogen bonding groups in proteins includes all the Coulombic interactions. J. Mol. Biol. 1995;248:361–373. doi: 10.1016/s0022-2836(95)80056-5. [DOI] [PubMed] [Google Scholar]
- 78.Mandell DJ, Kortemme T. Backbone flexibility in computational protein design. Curr. Opin. Biotechnol. 2009;20:420–428. doi: 10.1016/j.copbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 79.Monastyrskyy B, Fidelis K, Moult J, Tramontano A, Kryshtafovych A. Evaluation of disorder predictions in CASP9. Proteins. 2011;79(S10):107–118. doi: 10.1002/prot.23161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morozov AV, Kortemme T, Tsemekhman K, Baker D. Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations. Proc. Natl. Acad. Sci. USA. 2004;101:6946–6951. doi: 10.1073/pnas.0307578101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murphy GS, Mills JL, Miley MJ, Machius M, Szyperski T, Kuhlman B. Increasing sequence diversity with flexible backbone protein design: the complete redesign of a protein hydrophobic core. Structure. 2012;20:1086–1096. doi: 10.1016/j.str.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Offredi F, Dubail F, Kischel P, Sarinski K, Stern AS, et al. De novo backbone and sequence design of an idealized α/β-barrel protein: evidence of stable tertiary structure. J. Mol. Biol. 2003;325:163–174. doi: 10.1016/s0022-2836(02)01206-8. [DOI] [PubMed] [Google Scholar]
- 83.Paulini R, Muller K, Diederich F. Orthogonal multipolar interactions in structural chemistry and biology. Angew. Chem. Int. Ed. 2005;44:1788–1805. doi: 10.1002/anie.200462213. [DOI] [PubMed] [Google Scholar]
- 84.Pinto AL, Hellinga HW, Caradonna JP. Construction of a catalytically active iron superoxide dismutase by rational protein design. Proc. Natl. Acad. Sci. USA. 1997;94:5562–5567. doi: 10.1073/pnas.94.11.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pokala N, Handel TM. Review: protein design—where we were, where we are, where we’re going. J. Struct. Biol. 2001;134:269–281. doi: 10.1006/jsbi.2001.4349. [DOI] [PubMed] [Google Scholar]
- 86.Pokala N, Handel TM. Energy functions for protein design I: efficient and accurate continuum electrostatics and solvation. Protein Sci. 2004;13:925–936. doi: 10.1110/ps.03486104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pokala N, Handel TM. Energy functions for protein design: adjustment with protein-protein complex affinities, models for the unfolded state, and negative design of solubility and specificity. J. Mol. Biol. 2005;347:203–227. doi: 10.1016/j.jmb.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 88.Poole AM, Ranganathan R. Knowledge-based potentials in protein design. Curr. Opin. Struct. Biol. 2006;16:508–513. doi: 10.1016/j.sbi.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 89.Poso D, Sessions RB, Lorch M, Clarke AR. Progressive stabilization of intermediate and transition states in protein folding reactions by introducing surface hydrophobic residues. J. Biol. Chem. 2000;275:35723–35726. doi: 10.1074/jbc.M001747200. [DOI] [PubMed] [Google Scholar]
- 90.Privalov PL. Stability of proteins: small globular proteins. Adv. Protein Chem. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- 91.Quinn TP, Tweedy NB, Williams RW, Richardson JS, Richardson DC. Betadoublet: de novo design, synthesis, and characterization of a β-sandwich protein. Proc. Natl. Acad. Sci. USA. 1994;91:8747–8751. doi: 10.1073/pnas.91.19.8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Regan L, DeGrado WF. Characterization of a helical protein designed from first principles. Science. 1988;241:976–978. doi: 10.1126/science.3043666. [DOI] [PubMed] [Google Scholar]
- 93.Reina J, Lacroix E, Hobson SD, Fernandez-Ballester G, Rybin V, et al. Computer-aided design of a PDZ domain to recognize new target sequences. Nat. Struct. Biol. 2002;9:621–627. doi: 10.1038/nsb815. [DOI] [PubMed] [Google Scholar]
- 94.Rohl CA, Strauss CEM, Misura KMS, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 95.Romero P, Obradovic Z, Li XH, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins Struct. Funct. Genet. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 96.Rost B. Review: Protein secondary structure prediction continues to rise. J. Struct. Biol. 2001;134:204–218. doi: 10.1006/jsbi.2001.4336. [DOI] [PubMed] [Google Scholar]
- 97.Russ WP, Ranganathan R. Knowledge-based potential functions in protein design. Curr. Opin. Struct. Biol. 2002;12:447–452. doi: 10.1016/s0959-440x(02)00346-9. [DOI] [PubMed] [Google Scholar]
- 98.Sali A, Blundell TL. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 99.Samanta U, Bahadur RP, Chakrabarti P. Quantifying the accessible surface area of protein residues in their local environment. Protein Eng. 2002;15:659–667. doi: 10.1093/protein/15.8.659. [DOI] [PubMed] [Google Scholar]
- 100.Saunders CT, Baker D. Recapitulation of protein family divergence using flexible backbone protein design. J. Mol. Biol. 2005;346:631–644. doi: 10.1016/j.jmb.2004.11.062. [DOI] [PubMed] [Google Scholar]
- 101.Schindler T, Perl D, Graumann P, Sieber V, Marahiel MA, Schmid FX. Surface-exposed phenylalanines in the RNP1/RNP2 motif stabilize the cold-shock protein CspB from Bacillus subtilis. Proteins Struct. Funct. Genet. 1998;30:401–406. doi: 10.1002/(sici)1097-0134(19980301)30:4<401::aid-prot7>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 102.Schneider M, Fu X, Keating AE. X-ray versus NMR structures as templates for computational protein design. Proteins. 2009;77:97–110. doi: 10.1002/prot.22421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schreier B, Stumpp C, Wiesner S, Hocker B. Computational design of ligand binding is not a solved problem. Proc. Natl. Acad. Sci. USA. 2009;106:18491–18496. doi: 10.1073/pnas.0907950106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shah PS, Hom GK, Ross SA, Lassila JK, Crowhurst KA, Mayo SL. Full-sequence computational design and solution structure of a thermostable protein variant. J. Mol. Biol. 2007;372:1–6. doi: 10.1016/j.jmb.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 105.Shifman JM, Mayo SL. Modulating calmodulin binding specificity through computational protein design. J. Mol. Biol. 2002;323:417–423. doi: 10.1016/s0022-2836(02)00881-1. [DOI] [PubMed] [Google Scholar]
- 106.Siegel JB, Zanghellini A, Lovick HM, Kiss G, Lambert AR, et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Simons KT, Ruczinski I, Kooperberg C, Fox BA, Bystroff C, Baker D. Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins Struct. Funct. Genet. 1999;34:82–95. doi: 10.1002/(sici)1097-0134(19990101)34:1<82::aid-prot7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 108.Stordeur C, Dalluge R, Birkenmeier O, Wienk H, Rudolph R, et al. The NMR solution structure of the artificial protein M7 matches the computationally designed model. Proteins. 2008;72:1104–1107. doi: 10.1002/prot.22107. [DOI] [PubMed] [Google Scholar]
- 109.Suarez M, Jaramillo A. Challenges in the computational design of proteins. J. R. Soc. Interface. 2009;6:S477–S491. doi: 10.1098/rsif.2008.0508.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun S, Brem R, Chan HS, Dill KA. Designing amino acid sequences to fold with good hydrophobic cores. Protein Eng. 1995;8:1205–1213. doi: 10.1093/protein/8.12.1205. [DOI] [PubMed] [Google Scholar]
- 111.Taylor WR, Chelliah V, Hollup SM, MacDonald JT, Jonassen I. Probing the “dark matter” of protein fold space. Structure. 2009;17:1244–1252. doi: 10.1016/j.str.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 112.Tsai HH, Tsai CJ, Ma BY, Nussinov R. In silico protein design by combinatorial assembly of protein building blocks. Protein Sci. 2004;13:2753–2765. doi: 10.1110/ps.04774004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Virnau P, Mallam A, Jackson S. Structures and folding pathways of topologically knotted proteins. J. Phys. Condens. Matter. 2011;23:033101. doi: 10.1088/0953-8984/23/3/033101. [DOI] [PubMed] [Google Scholar]
- 114.Vizcarra CL, Mayo SL. Electrostatics in computational protein design. Curr. Opin. Chem. Biol. 2005;9:622–626. doi: 10.1016/j.cbpa.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 115.Walsh ST, Cheng H, Bryson JW, Roder H, DeGrado WF. Solution structure and dynamics of a de novo designed three-helix bundle protein. Proc. Natl. Acad. Sci. USA. 1999;96:5486–5491. doi: 10.1073/pnas.96.10.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wernisch L, Hery S, Wodak SJ. Automatic protein design with all atom force-fields by exact and heuristic optimization. J. Mol. Biol. 2000;301:713–736. doi: 10.1006/jmbi.2000.3984. [DOI] [PubMed] [Google Scholar]
- 117.Wootton JC, Federhen S. Statistics of local complexity in amino acid sequences and sequence databases. Comput. Chem. 1993;17:149–163. [Google Scholar]
- 118.Yang Y, Faraggi E, Zhao H, Zhou Y. Improving protein fold recognition and template-based modeling by employing probabilistic-based matching between predicted one-dimensional structural properties of the query and corresponding native properties of templates. Bioinformatics. 2011;27:2076–2082. doi: 10.1093/bioinformatics/btr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang YD, Zhou Y. Specific interactions for ab initio folding of protein terminal regions with secondary structures. Proteins. 2008;72:793–803. doi: 10.1002/prot.21968. [DOI] [PubMed] [Google Scholar]
- 120.Zhang T, Faraggi E, Xue B, Dunker AK, Uversky VN, Zhou Y. SPINE-D: accurate prediction of short and long disordered regions by a single neural-network based method. J. Biomol. Struct. Dyn. 2012;28:799–813. doi: 10.1080/073911012010525022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou H, Zhou Y. Fold recognition by combining sequence profiles derived from evolution and from depth-dependent structural alignment of fragments. Proteins. 2005;58:321–328. doi: 10.1002/prot.20308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhou HY, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhou Y, Zhou HY, Zhang C, Liu S. What is a desirable statistical energy function for proteins and how can it be obtained? Cell Biochem. Biophys. 2006;46:165–174. doi: 10.1385/cbb:46:2:165. [DOI] [PubMed] [Google Scholar]