Figure 3. Phylogenetic tree based on the amino acid sequences of TDs from 82 representative species.
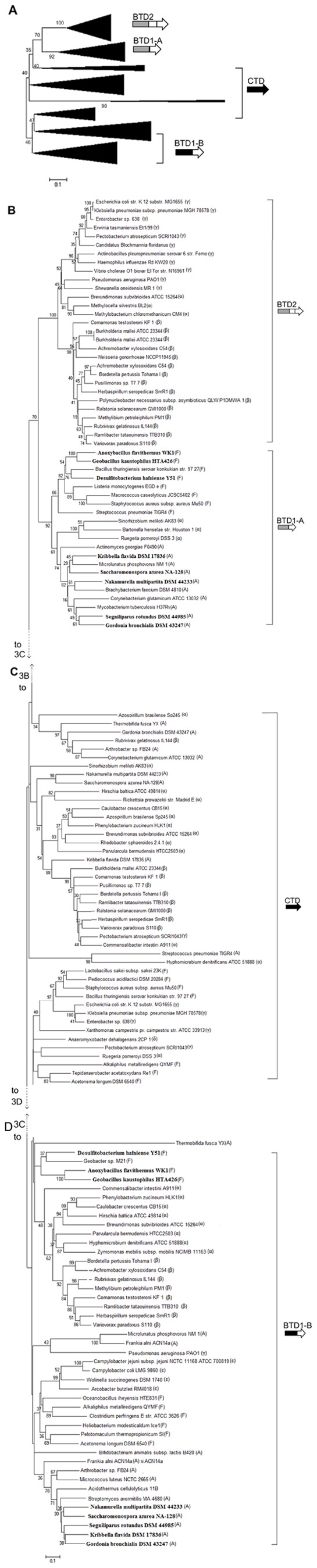
Genes encoding the enzymes are represented by arrows. The overall structure of the phylogenetic tree is shown in A. Because it is too big to show in a single page, the detail structure of the phylogenetic tree is divided into three panels (B, C and D). The connecting point of the tree segments in the three panels is marked with a broken line. The strains shown in bold contain both genes encoding for BTD1-A and BTD1-B. α, β, δ, ε, γ, F and A indicate α-proteobacteria, β-proteobacteria, δ-proteobacteria, ε-proteobacteria, γ-proteobacteria, Firmicutes and Actinobacteria, respectively. The tree was constructed with the MEGA 5 software using the neighbor-joining method and 1000 bootstrap replicates.
