Abstract
Kar3Cik1 is a heterodimeric kinesin-14 from Saccharomyces cerevisiae involved in spindle formation during mitosis and karyogamy in mating cells. Kar3 represents a canonical kinesin motor domain that interacts with microtubules under the control of ATP-hydrolysis. In vivo, the localization and function of Kar3 is differentially regulated by its interacting stoichiometrically with either Cik1 or Vik1, two closely related motor homology domains that lack the nucleotide-binding site. Indeed, Vik1 structurally resembles the core of a kinesin head. Despite being closely related, Kar3Cik1 and Kar3Vik1 are each responsible for a distinct set of functions in vivo and also display different biochemical behavior in vitro. To determine a structural basis for their distinct functional abilities, we used cryo-electron microscopy and helical reconstruction to investigate the 3-D structure of Kar3Cik1 complexed to microtubules in various nucleotide states and compared our 3-D data of Kar3Cik1 with that of Kar3Vik1 and the homodimeric kinesin-14 Ncd from Drosophila melanogaster. Due to the lack of an X-ray crystal structure of the Cik1 motor homology domain, we predicted the structure of this Cik1 domain based on sequence similarity to its relatives Vik1, Kar3 and Ncd. By molecular docking into our 3-D maps, we produced a detailed near-atomic model of Kar3Cik1 complexed to microtubules in two distinct nucleotide states, a nucleotide-free state and an ATP-bound state. Our data show that despite their functional differences, heterodimeric Kar3Cik1 and Kar3Vik1 and homodimeric Ncd, all share striking structural similarities at distinct nucleotide states indicating a common mechanistic theme within the kinesin-14 family.
Keywords: Kinesin-14, Kar3Cik1, Microtubules, Helical 3-D analysis, Molecular docking, Cryo-electron microscopy
Introduction
Microtubule-based molecular motors of the kinesin superfamily are classified into fourteen different classes (Lawrence et al., 2004). Here we focus on the members of the kinesin-14 family, which, according to current knowledge are the only kinesins that are minus-end directed and form their motor domain at the C-terminal end of the polypeptide chain (Ncd: McDonald & Goldstein, 1990; Kar3Vik1: Endow et al., 1994; XCTK2: Walczak et al., 1997; HSET: Mountain et al., 1999). All of the kinesin-14’s that have been studied so far were found to be non-processive retrograde directed motors (Walker et al., 1990; Case et al., 1997; de Castro et al., 2000; Foster and Gilbert, 2000; Fink et al., 2009). Kinesin-14s contribute to an inward directed force towards the minus-end of the microtubules (Saunders & Hoyt 1992; Sharp et al., 1999). The most extensively studied kinesin-14 is Ncd from Drosophila melanogaster (McDonald et al., 1990; Endow et al., 1990, for structure-function investigations see: Hoenger et al., 1995; Sosa et al., 1997; Sablin et al., 1998; Wendt et al., 2002; Endres et al., 2006; Fink et al., 2009). Ncd is required for proper chromosome distribution in meiosis and early mitosis and has been implicated in having roles in bipolar spindle assembly and its maintenance as well as spindle pole formation (Endow et al., 1994; Oladipo et al., 2007). Ncd uses its C-terminal motor domains and N-terminal non-motor microtubule binding domains (Karabay & Walker, 1999; Wendt et al., 2003) to crosslink antiparallel microtubules within the spindle midzone and parallel microtubules at the spindle poles.
Of the six kinesins found in S. cerevisiae, one of them, Kar3, belongs to the kinesin-14 family. Kar3, like all other kinesin-14s contains a C-terminal motor-head domain. In combination with an N-terminal non-motor microtubule binding site it crosslinks microtubules during karyogamy and mitosis (Meluh & Rose 1990; Rose 1996). Kar3 is essential for yeast nuclear fusion during mating, but it is apparently not essential for mitotic growth (Meluh & Rose 1990; reviewed in: Winey & Bloom, 2012). The structure of the monomeric Kar3 motor domain has been solved to atomic detail by X-ray crystallography (Gulick et al., 1998) and shares most structural elements with other kinesin motor domains.
Kar3 function in vivo is regulated by its capacity to selectively heterodimerize with either of two motor-homology domains called Vik1 and Cik1 (Manning et al., 1999). While Vik1 and Cik1 have a relatively high level of sequence similarity (24% identity, 37% homology: Manning et al., 1999), their localization during the yeast life cycle and genetic analysis demonstrated that Kar3Vik1 and Kar3Cik1 exhibit remarkably different properties and are responsible for carrying out distinct sets of functions (Manning et al., 1999). Kar3Vik1 is only expressed during mitosis where it localizes predominantly to the poles of the mitotic spindle and likely contributes to spindle stabilization by crosslinking and focusing the minus-ends of parallel microtubules as proposed for other kinein-14 motors (Manning et al., 1999, Allingham et al., 2007). An EM structure (Cope et al., 2010; Cope et al., in press), as well as an X-ray crystal structure (Rank et al., 2012) of a truncated heterodimeric Kar3Vik1 were published recently. In contrast to Kar3Vik1, Kar3Cik1 crosslinks anti-parallel interpolar MTs in the overlap zone, providing stabilization and controlling the spindle mid-region geometry (Gardner et al., 2008). Upon pheromone treatment, Kar3Cik1 localizes to cytoplasmic microtubules (Page et al., 1994) to facilitate karyogamy by shortening microtubules, pulling the two nuclei together for nuclear fusion (Page & Snyder 1992, Page et al., 1994; Maddox et al., 2003). In support of this, Kar3Cik1 has been shown to have robust depolymerizing activity in vitro predominantly from microtubule plus-ends (Sproul et al., 2005). Conversely, Kar3Vik1 only gradually and non-specifically depolymerizes MTs from both the plus and minus ends in vitro, similar to the weak depolymerizing activity reported for Ncd (Sproul et al., 2005; Allingham et al., 2007). Though Cik1 is not essential for mitotic growth (Page et al., 1994; Manning et al., 1999), Kar3Cik1 localizes predominantly to the interpolar MTs at the midzone of the mitotic spindle where it is thought to crosslink antiparallel microtubules and provide an inward force during spindle assembly (Meluh & Rose 1990; Page et al., 1994, Gardner et al., 2008).
The functional differences between Kar3Vik1 and Kar3Cik1 despite being closely related, prompted us to investigate the microtubule-bound structure of Kar3Cik1, particularly in comparison with Kar3Vik1 in an attempt to obtain insight into how these two complexes are uniquely suited to carry out their respective functions. Here we have used electron microscopy to study the conformation of a heterodimeric Kar3Cik1 motor domain construct in the presence of ADP and AMPPNP (mimicking an ATP state), as well as after apyrase treatment to generate the nucleotide-free state. We also report an intermediate resolution 3-D structure of Kar3Cik1 complexed to microtubules in the nucleotide-free and ATP states. Our 3-D structures were obtained by cryo-electron microscopy (cryo-EM) and 3-D image reconstruction based on the helical symmetry of microtubules composed of 15 protofilaments (Arnal et al., 1996; reviewed in Hoenger and Gross, 2008). Our results show that Kar3Cik1’s coiled-coil stalk rotates ~65° upon uptake of AMPPNP representing the powerstroke that Kar3Cik1 uses to move toward the microtubule minus-end. By comparing the structures of Kar3Cik1, Kar3Vik1, and homodimeric Ncd (Hirose et al., 1996; Sosa et al., 1997; Wendt et al., 2002) we found that all these kinesin-14 motors, whether heterodimeric or homodimeric in nature, show almost identical 3-D microtubule-bound conformations in both the nucleotide-free and ATP-states. Hence, despite some differences between the densities of the Vik1 and Cik1 domains, both complexes show a common theme for a powerstroke mechanism, highly reminiscent of that described for Ncd (Wendt et al., 2002, Endres et al., 2006). However, while movements of the structural elements of Kar3Cik1 and Kar3Vik1 are similar, here we also provide evidence that their microtubule-binding patterns differ in that Kar3Vik1 binds to microtubules in a highly cooperative manner while Kar3Cik1 binds stochastically. While the basis for cooperative binding by Kar3Vik1 is not yet known, this difference may partially explain how Kar3Cik1 has adapted to its role as a microtubule depolymerizing factor where other motors in close proximity may be a hindrance for MT shortening while Kar3Vik1 works in concert by recruiting additional motors to the spindle poles to facilitate spindle assembly and stabilization.
Results
High-affinity Kar3Cik1 motor-microtubule binding conformations
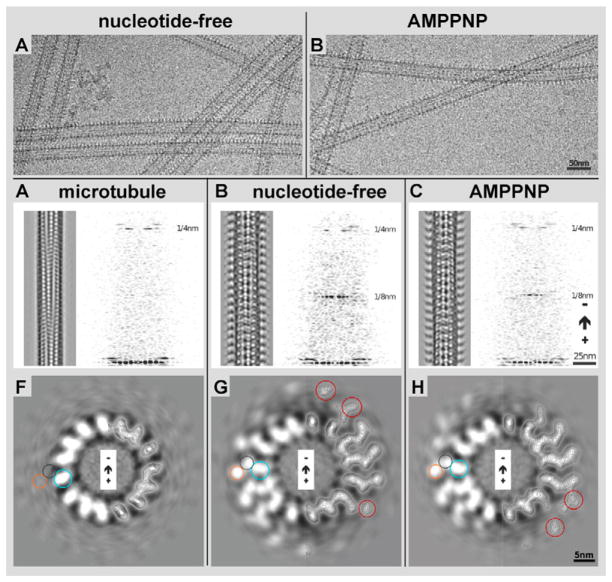
We have analyzed the 3-D microtubule-binding conformation of Kar3Cik1 by cryo-EM, helical 3-D image reconstruction, and molecular docking (see Figs. 1, 2 & 4). Micrographs of frozen-hydrated motor-microtubule complexes were screened for fully decorated 15 protofilament microtubules that were then reconstructed in 3-D by the PHOELIX/SUPRIM image-processing package (Whittaker et al., 1995; Schroeter & Bretaudiere, 1996). Polymerized, taxol-stabilized microtubules were mixed at a molar ratio of 1:6 αβ-tubulin to Kar3Cik1 heterodimers and incubated with either ADP (see Fig. 3), apyrase to generate the nucleotide-free state, or AMPPNP, a non-hydrolyzable ATP analog to generate an ATP-bound state (Fig. 1A & B). Excess ADP (1 mM) generates a weak microtubule affinity in Kar3Cik1, which at our experimental conditions produced only sparsely decorated microtubules unsuitable for helical 3-D analysis. Instead, the ADP state was further analyzed by high-resolution surface metal shadowing (see Fig. 3) discussed below. All other nucleotide states produced high-affinity binding. Nucleotide-free conditions, or the presence of excess AMPPNP (1 mM) revealed fully decorated microtubules with a binding stoichiometry of one Kar3Cik1 heterodimer on each tubulin dimer. The degree of decoration was estimated from diffraction patterns and the power of the characteristic 8 nm and 4 nm layerlines (Fig. 1C–E). On raw-data images as shown in Figure 1A and B, structural differences between the two nucleotide states are essentially invisible due to the inherently low signal to noise ratio of cryo-micrographs (Fig. 1A & B), though the micrographs can be enhanced with Fourier-filtering to reveal distinct conformational changes (left panels in Fig. 1C, D, & E). After 3-D reconstruction and averaging, density differences appear clearly on 5 nm thick slices through the reconstructed 3-D density maps (see Fig. 1G & H, and Fig. 6) as well as on surface rendered volumes (iso-surface maps: Figs. 2, 4 & 7).
Figure 1. 3-D helical reconstruction of decorated and undecorated microtubules.
Microtubule/Kar3Cik1 solutions were incubated with apyrase (A) or AMPPNP (B) to generate the nucleotide-free and ATP states respectively. In both states, Kar3Cik1 heterodimers can be seen decorating the microtubules although structural differences between the states cannot be detected. C) 3D helical reconstruction of a plain microtubule. The same result was obtained by incubating Kar3Cik1 motors with ADP prior to addition to microtubules, but where no decoration was detected. A Fourier transform reveals only the 4 nm layer line, which corresponds to the tubulin repeats, illustrating only sparse decoration with motor complexes. D) Reconstruction of a Kar3Cik1 decorated microtubule in the nucleotide-free state. Two distinct densities are visible at each motor subunit corresponding to Kar3 and Cik1. The Fourier transform reveals a strong 4 nm layer line (tubulin repeats) as well as an 8 nm layer line that corresponds to the motor repeat every 8 nm along the microtubule. E) Reconstruction of the ATP state using the non-hydrolyzable ATP analog AMPPNP. The microtubule is fully decorated with Kar3Cik1 as in (D) illustrated by the strong 4 nm and 8 nm layer lines. Corresponding cross-sectional views of the reconstructed microtubules are shown in F) undecorated microtubule, G) nucleotide-free state, and H) AMPPNP state. On the right half of each image, contour lines are shown depicting densities observed from tubulin and Kar3Cik1. Circles corresponding to the position of tubulin (blue), Kar3 (purple), and Cik1 (orange) are shown as well as red circles identifying differences in density between the nucleotide-free and AMPPNP states
Figure 2. Isosurface rendering of density maps obtained by helical reconstruction.
A) Isosurface of a naked 15-protofilament microtubule showing the orientation of tubulin (turquoise). B) The nucleotide-free Kar3Cik1 is in complex with the microtubule. It appears that Kar3 (grey / black rings) is in contact with the microtubule while Cik1 (orange / red rings) is oriented away from the microtubule. The coiled coil stalk (light blue / blue rings) connecting Kar3 and Cik1 is pointed toward the microtubule plus end. C) In the ATP state Kar3Cik1 is also in complex with the microtubule with Kar3 in contact with the microtubule and Cik1 oriented away. Interestingly, there is a change in Kar3Cik1’s conformation showing a ~65° rotation of the stalk that causes it to point toward the minus end of the microtubule (black arrows). Other subtle differences can be seen when comparing the orange globular region corresponding to Cik1 (red rings). A cross-sectional view of D) a naked microtubule, E) the nucleotide-free, and F) ATP states shows how Kar3Cik1 (grey and orange) is in contact with tubulin (turquoise).
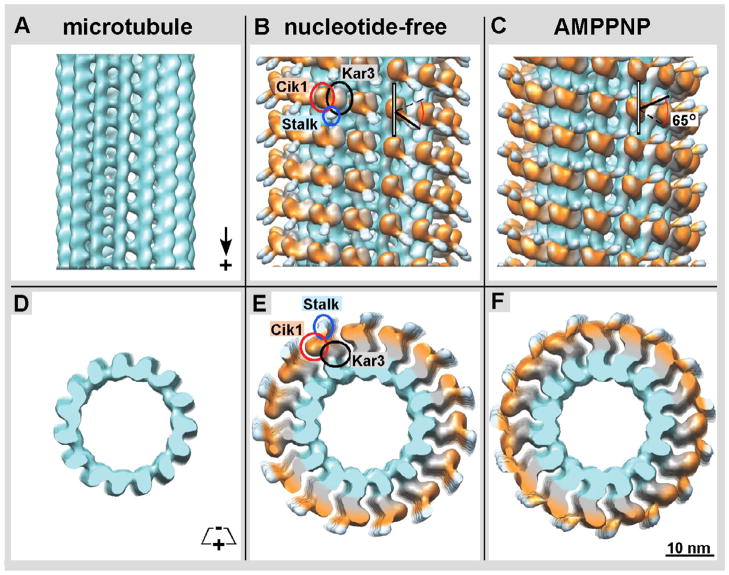
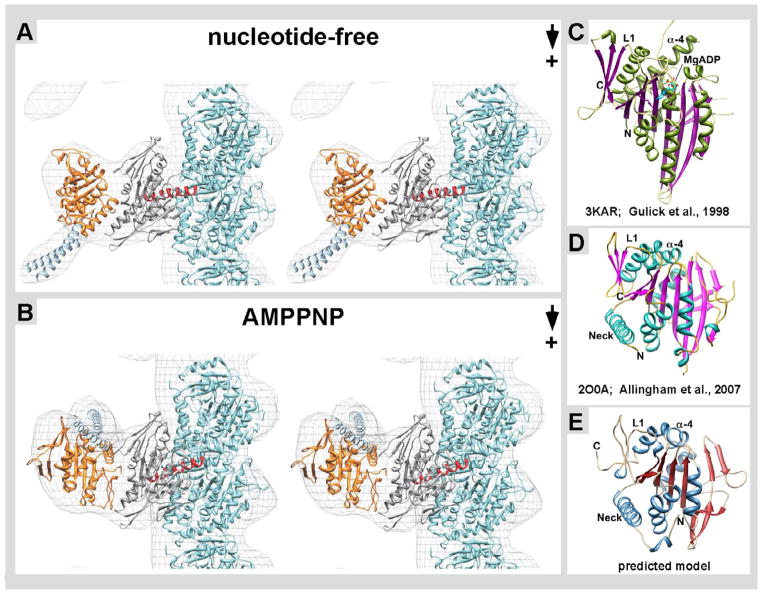
Figure 4. Docking of near-atomic structures into cryo-EM derived isosurfaces.
A) In the nucleotide-free state, the structure of tubulin (turquoise, PDB: 1JFF) is shown and the Kar3 structure (grey, PDB: 3KAR) is positioned in the globular region in contact with β-tubulin. Kar3’s helix α-4 (red) is at the interface of where Kar3 contacts the microtubule. As there is currently no known structure for Cik1, a predicted structure was obtained using Phyre 2 (Kelley & Sternberg 2009). This predicted Cik1 structure (orange) is positioned in the globular region oriented away from the microtubule. The structure of the GCN4 leucine zipper (light blue, PDB: 2ZTA) is docked into the stalk region. In the nucleotide-free state the stalk is pointing toward the plus end of the microtubule. B) In the ATP state, there is a conformational change in Kar3Cik1 that leads to a ~65° rotation resulting in the stalk pointing toward the minus end of the microtubule. Phyre2 used structural information from C) Kar3, D) Vik1, and other proteins to generate the predicted model of E) Cik1.
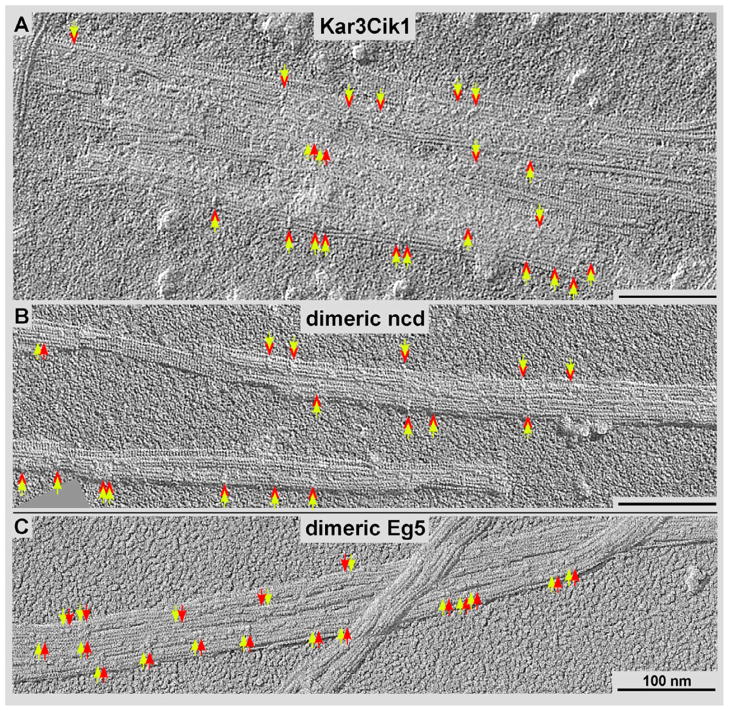
Figure 3. High-resolution surface metal shadowing of kinesin motors.
Kinesin motors bound to ADP were incubated with microtubules at sub-stoichiometric levels and shadowed with tantalum/tungsten. Dimeric motor constructs are identified by a yellow arrow pairs in A) Kar3Cik1, B) dimeric Ncd, and C) dimeric Eg5 (a kinesin-5). The kinesin-14 motors appear to bind sparsely along the microtubules and span adjacent protofilaments (red relative to yellow arrows in A and B) while dimeric kinesin-5s appear to bind to adjacent tubulin subunits along the same protofilament (red relative to yellow arrows in C).
Figure 6. Structural Comparison of Kar3Cik1 and Kar3Vik1.
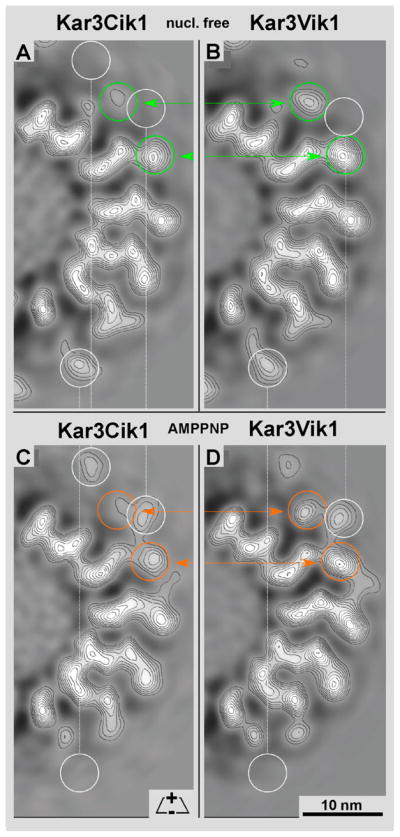
By comparing the density map of a single 3.8 nm slice of A) Kar3Cik1 and B) Kar3Vik1 in the nucleotide-free state, a difference in the density of the motors is detected (green circles). Cik1 appears to have less density present than Vik1, which may suggest that Cik1 is more mobile than Vik1. The same observation is seen when comparing C) Kar3Cik1 and D) Kar3Vik1 in the ATP state. Cik1 again seems to have less density (and thus perhaps more flexibility) than Vik1 (orange circles). White circles show differences in Kar3Cik1 and Kar3Vik1 between their own nucleotide-free and ATP states. Since these 15-protofilament microtubules exhibit a dominant 2-start short-pitched helix (Bessel order -2), a thin slice (~4nm) of a half cross-section, as shown here, represents the complete density distribution of a single motor from these maps.
Figure 7. 3-D structural comparison of Kar3Cik1 and Kar3Vik1.
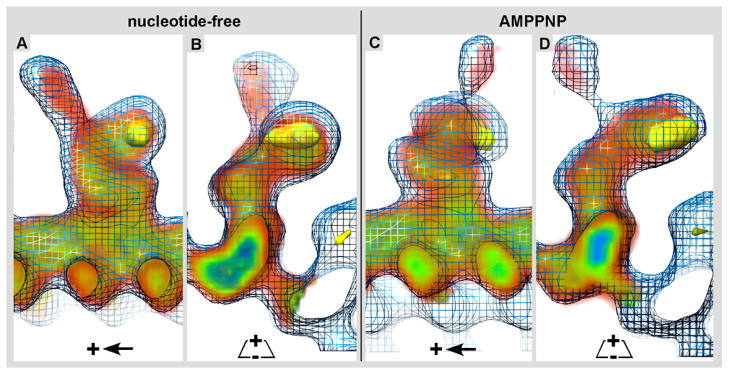
The 3-D electron density map of Kar3Cik1 was docked into the isosurface mesh representation of the 3-D electron density map of Kar3Vik1. Longitudinal, A) and cross-sectional B) views of the nucleotide-free state show that Kar3Cik1 docks into the Kar3Vik1 map extremely well demonstrating that Kar3Cik1 adopts the same pre-powerstroke configuration as Kar3Vik1. Similarly, longitudinal C) and cross-sectional D) views of the AMPPNP state confirm that Kar3Cik1 fits excellently into the Kar3Vik1 structure showing a nearly identical post-powerstroke position following uptake of ATP. Difference mapping (yellow) reveals locations of density differences between Kar3Vik1 and Kar3Cik1 with a significance of >95%. Interestingly, these results show that overall, Kar3Cik1 and Kar3Vik1 appear structurally identical at this resolution, and it seems that they utilize the same mechanism of movement to perform different functions in the cell. Wire mesh: Kar3Vik1; red/green/blue diffuse density: Kar3Cik1; solid yellow density: difference map at >95% significance.
The iso-surface representations of our 3-D reconstructions enclose roughly 90–95% of the actual protein mass, to cancel out excess noise-related densities. Iso-surfaces do not reveal any inner density variations as shown in cross-sections through the maps (Figs. 1F–H & 6), but they provide a comprehensive overview of the entire structure and nicely illustrate large conformational changes. In both, the nucleotide-free and AMPPNP states we find one domain of the Kar3Cik1 heterodimer to contact the microtubule surface, while the second domain protrudes outwards without touching the microtubule surface. The shapes of the domains and their 3-D configuration are very similar to Kar3Vik1 (inner density in Fig. 7) and only the plus-end oriented region of the Cik1 density shows some variation to Vik1 (yellow volume in Fig. 7). In addition, the predicted high-resolution structure of the Cik1 motor homology domain (Fig. 4E) strongly resembles that of Vik1 (Fig. 4D; Allingham et al., 2007; Rank et al., 2012) while both are quite different from the Kar3 motor domain (Fig. 4C; Gulick et al., 1998; Rank et al., 2012). In our previous Kar3Vik1-microtubule reconstructions the two domains were unambiguously identified by site-directed maleimide-gold labeling (Cope et al., in press). Hence, based on structural similarities, especially in the Kar3 domain we can safely state that analogous to Kar3Vik1 it is again the Kar3 motor domain of Kar3Cik1 that maintains contact with the microtubule surface while the Cik1 motor homology domain protrudes outwards (Fig. 1G & H, Figs. 2 & 4).
Reminiscent of dimeric Ncd (Wendt et al., 2002; Endres et al., 2006) and Kar3Vik1 (Fig. 7; Rank et al., 2012; Cope et al., in press), a most striking conformational change occurs upon AMPPNP uptake into the empty nucleotide pocket of the Kar3 motor domain. During the transition from the nucleotide-free to AMPPNP-bound state, the overall microtubule-binding geometry of Kar3 remains unchanged. However, the Cik1 head domain rotates substantially (Fig. 2B & C). This rotation consists of a 65-degree swing of Kar3Cik1’s truncated heterodimeric stalk (Fig. 2B, C, E, F) that is held together by an engineered GCN4 dimerization motif (see: Rank et al., 2012). The stalk region that stabilizes the heterodimer can be seen as a relatively weak, but nevertheless well defined elongated density which is pointing toward the plus end of the microtubule. The cross-sectional views shown in Figures 1 and 6 allow us to study in great detail the inner density distributions in the 3-D data and how Kar3Cik1 interacts with the tubulin subunits of the microtubule. The binding geometry of Kar3 motor head to tubulin is very similar to monomeric kinesins constructs studied with this method (e.g. see Sosa et al., 1997 (Ncd) Hoenger et al., 2000 (kinesin-1); Hirose et al., 2006 (Kar3)). The Cik1 motor homology domain adopts a very similar conformation as the second motor heads in dimeric Ncd constructs (Sosa et al., 1997; Wendt et al., 2002; Endres et al., 2006) and Kar3Vik1 constructs (Cope et al., 2010; Rank et al., 2012; Cope et al., in press). It is tethered to the Kar3 motor domain, protrudes outwards and does not touch the microtubule surface.
Low-affinity Kar3Cik1-microtubule binding patterns
As for most kinesin motor domains, ADP-bound Kar3 motor domains maintain a low microtubule-affinity. Complete microtubule decoration would be required for a quantitative analysis by helical averaging but this is often very difficult to achieve. 3-D image analysis of frozen-hydrated complexes are usually not conclusive and tomographic reconstructions, unless volume averaging could be applied are typically too noisy (see Cope et al., 2010) for quantitative image analysis. Nevertheless, ADP reveals sparse decoration of Kar3Cik1 complexes that is easily detectable with high-resolution, unidirectional surface metal shadowing (Fig. 3A). The power of this method is its focus on surface structures through an almost complete elimination of inner density contributions, which dominate cryo-EM projections (e.g. see Fig. 1A & B, and Fig. 6). Figure 3 compares the surface-binding patterns of ADP-Kar3Cik1 (Fig. 3A) with that of ADP-Ncd (Fig. 3B), which both show striking similarities to ADP-Kar3Vik1 (Rank et al., 2012). In all three cases it appears as if all of these kinesin-14 dimers adopt a dimeric binding configuration that spans two adjacent protofilaments. While this configuration is not always obvious, it becomes clear that this binding configuration is very different from motors such as dimeric kinesin-1 (Hoenger et al., 2000) or kinesin-5 (Fig. 3C; see Krzysiak et al., 2006) that bind predominantly with both motors to two adjacent tubulin dimers along the same protofilament (Fig. 3C). Within the same context we observed very little microtubule binding affinity of both monomeric Cik1 (this work) and Vik1 constructs (see Cope et al., 2013).
Prediction of the Cik1 domain fold
Currently there are two independent atomic X-ray structures available for Vik1 from crystals made of a monomeric construct (Allingham et al., 2007) as well as the heterodimeric complex of Kar3Vik1 (Rank et al., 2012). However, so far no such structure is available for Cik1, though our EM results strongly suggest that the overall globular shape is very similar to that of Vik1. To improve the accuracy of our Kar3Cik1 dimer-docking attempts (Fig. 4A & B) and to determine whether Cik1 would adopt similar secondary structure elements as Vik1 (Fig. 4D) we predicted the Cik1 structure (Fig. 4E) with the help of the protein structure prediction software Phyre2 (Kelley & Sternberg, 2009). This algorithm generates a 3-D tertiary structure model based on protein sequence, fold recognition, and by comparisons to 3-D structures of similar proteins. In our attempts Vik1 (25% sequence identity to Cik1) (Allingham et al., 2007; PBD: 2O0A) and Ncd (Sablin et al., 1998; PDB: 2NCD) were the most important contributors to the predicted Cik1 structure. The predicted model (Fig. 4E) is composed of residues Asn344 – Asp594, which includes part of Cik1’s neck-helix that dimerizes with Kar3 (Fig. 4C). The resulting structure shows many similarities with Vik1 (Fig. 4D) and some of the folding patterns observed in kinesin motor domains that have been discussed in detail by Allingham et al. (2007). These are in particular the conserved arrangement of three inner β-sheets and most of the surrounding helices. Overall the predicted Cik1 structure and folding pattern strongly resembles that of Vik1, and at a resolution of ~2.5 nm as in the EM maps, the differences are marginal.
Docking X-ray crystal structures into the 3-D EM scaffold
Despite numerous attempts, so far no kinesin-motor domain has been crystalized in complex with the αβ-tubulin dimer and solved to near-atomic resolution. Hence, the structure of the motor-tubulin interface has been modeled by docking the atomic-resolution X-ray crystal structures of various kinesin motor domains and the electron crystal structures of the αβ-tubulin dimer (Nogales et al., 1998; Löwe et. al., 2001; PDB: 1JFF) into cryo-EM maps of motor-microtubule complexes (for monomeric Kar3 see: Hirose et al., 2006; Kar3Vik1: Rank et al., 2012 and Cope et al., in press; Ncd: Sosa et al., 1997; Wendt et al., 2002; Endres et al., 2006).
Figure 4A and B show the docking of X-ray crystal structures of monomeric Kar3 (grey; Gulick et al., 1998; PDB: 3KAR) together with our predicted model of Cik1 (orange in Fig. 4A & B; see also Fig. 4E). In Figure 4A, the near-atomic resolution structures of αβ-tubulin, Kar3, Cik1 and the stalk including the GCN4 leucine zipper sequence used to initialize dimerization (O’Shea et al., 1991; PDB: 1ZTA; colored light blue in Fig. 4), were docked into the nucleotide-free state. The refined tubulin dimer structure (Löwe et. al., 2001) was docked into the density corresponding to the microtubule. The structure of Kar3 (Gulick et al., 1998) was docked into the globular region that is in contact with the microtubule (see also Hirose et al., 2006). Helix α-4 of Kar3 (shown in red) forms part of the motor’s so-called switch-II region (Vale & Milligan, 2000) and was positioned according to Hirose et al. (2006) in contact with the highly negatively charged C-terminal helix-12 and tail region of β-tubulin.
In both the nucleotide-free and ATP states, the predicted Cik1 structure fits the EM structure with remarkable precision (Fig. 4A & B). As discussed above (Fig. 2), there are significant rearrangements from the nucleotide-free configuration that occur upon AMPPNP uptake. Most importantly, the AMPPNP-bound conformation has the stalk pointing toward the minus end of the microtubule. This result shows that Kar3Cik1 uses a stalk rotation for minus-end directed movement whose angle and timing in the nucleotide-hydrolysis cycle strongly resemble the powerstroke shown for Kar3Vik1 (Rank et al., 2012; Cope et al., in press) as well as that of homodimeric Ncd (Wendt et al., 2002; Endres et al., 2006). Figure 4A & B shows stereo pairs of the two distinct conformations that allow the docked structures to be viewed in 3-D. The powerstroke that occurs between the nucleotide-free and AMPPNP states (Figs. 2 & 4) implies that the Cik1 motor homology domain undergoes a ~65 degree rotation around a pivot point near the C-terminus of the stalk dimerization motif, at the end of the neck. The stalk region has to rotate as shown to fit into the density map, but also to avoid steric clashes between the Kar3 and Cik1 domains.
Structural Comparisons between the nucleotide-dependent 3-D conformations of Kar3Cik1 and Kar3Vik1
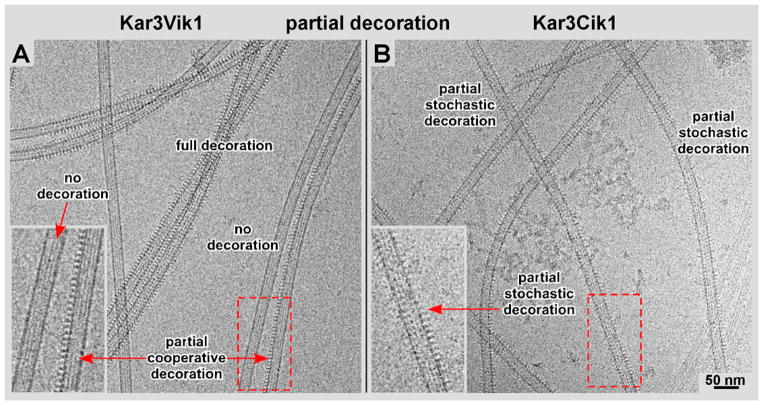
Vik1 and Cik1 are both motor homology domains in S. cerevisiae that lack nucleotide-binding sites, but serve to regulate Kar3’s localization and function by differentially pairing with Kar3 to form a functional kinesin-14 complex. Detailed biochemical studies have been carried out on both heterodimeric complexes (Allingham et al., 2007, Chen et al., 2012 (both: Kar3Vik1); Chen et al., 2011 (Kar3Cik1)), but to date no direct structural comparisons are available. In our hands, the microtubule binding properties of both motor complexes with regard to nucleotide state have been rather similar. Both complexes show weak microtubule binding affinity in the presence of ADP, but very strong binding affinity in the absence of nucleotide and in the presence of AMPPNP. In both strong binding affinity conditions, Kar3Vik1 binds to microtubules with a striking cooperative behavior (Fig. 5A; Cope et al., 2010), very much like Ncd (see Fig. 2 in Wendt et al., 2002). However, for Kar3Cik1 this cooperative binding pattern is much less pronounced (Fig. 5B). Microtubule binding is more stochastic and resembles that of monomeric Kar3 (Sproul et al., 2005; Allingham et al., 2007; Cope et al., in press) or other kinesins such as dimeric kinesin-1 (Hoenger et al., 2000) or kinesin-5 (Eg5: Krzysiak et al., 2006; see also Fig. 3C). The cooperative binding of Kar3Vik1 is best seen on projections of the outer microtubule walls, which are sometimes fully decorated and sometimes completely empty (inset and arrows in Fig. 5A). Some microtubules appear to be fully decorated while others are almost entirely free of motors. In contrast, the outer walls of microtubules decorated with Kar3Cik1 show scattered densities with numerous empty spots between the motors (inset and arrows in Fig. 5B), and decoration is much less clustered than with Kar3Vik1.
Figure 5. Kar3Vik1/Kar3Cik1 binding cooperativity.
Kar3Vik1 appears to bind more cooperatively than Kar3Cik1. A) Kar3Vik1 exhibits high cooperativity. Microtubules that are completely decorated, partially decorated, and undecorated can be seen in the same image area (see also: Cope et al., 2010). B) In contrast, Kar3Cik1 exhibits a more stochastic pattern, illustrated by scattered motor binding with numerous gaps appearing in between the motors. The difference in the Kar3Vik1 and Kar3Cik1 binding patterns is most easily visualized along the sidewalls of the microtubule projections (insets in A and B).
A thorough comparison between the reconstructions of Kar3Cik1 (this paper) and Kar3Vik1 (Cope et al., in press) in both the nucleotide-free (Fig. 6A & B; Fig. 7A & B) as well as the AMPPNP states (Fig. 6C & D; Fig. 7C & D) revealed many similarities. For the most part, their iso-surfaces are almost identical (Fig. 7) featuring only minor differences that are best seen on cross-sections through the 3-D densities (Fig. 6). Horizontal comparisons marked with colored rings highlight the most notable differences between Kar3Cik1 (Fig. 6A & C) and Kar3Vik1 (Fig. 6B & D). Vertical comparisons (white rings) show differences within each complex caused by the nucleotide state. The side-by-side comparison in Figure 6 reveals some positional shifts within the outermost domains that are formed by Cik1 and Vik1 respectively (arrows in Fig. 6). The conformational differences between Kar3Vik1 and Kar3Cik1 have been analyzed by statistical difference mapping (Fig. 7) based on a student’s t-test (Milligan & Flicker, 1987). The wireframe volume in Figure 7 represents Kar3Vik1, the red/green/blue densities are Kar3Cik1, and the solid yellow density marks the location of a difference volume representing a range of 99% significance. Despite the apparent low resolution of ~2.5 nm in these maps the positioning of mass centers with respect to each other between two aligned and normalized 3-D datasets can be assessed with sub-nanometer precision, resulting in difference maps as shown in Figure 7.
Discussion
Within the kinesin superfamily (for a unified nomenclature see: Lawrence et al., 2004), kinesin-14 members have distinct structural and functional properties that render them quite different from other kinesin families. These unique properties include their direction of movement towards the minus-end of microtubules, and their reversed building plan that places the motor domain at the C-terminal end of the polypeptide chain (Walker et al., 1990; McDonald et al., 1990; Endow et al., 1994; Mountain et al., 1999). In addition, all kinesin-14’s studied thus far are non-processive motors (Case et al., 1997; de Castro et al., 2000; Foster and Gilbert, 2000; Fink et al., 2009), and for all of them the cargo-binding site interacts with a second microtubule, but in a non-motor like fashion and without the control of nucleotide state (Karabay and Walker, 1999; Wendt et al., 2003). Hence, unlike kinesin-1 members or other highly processive kinesins, kinesin-14s would be very ineffective motors for long-distance transport. Instead, kinesin-14’s act in the densely packed microtubule arrangement of a bipolar spindle where they maintain the spindle arrangement and dynamics (Fink et al., 2009; reviewed in Peterman & Scholey, 2009).
The close relationship between the budding yeast (S. cerevisiae) Kar3Cik1 and Kar3Vik1 (recently reviewed in: Winey & Bloom, 2012), invites a careful exploration into the structural and functional differences between these two related heterodimers. How are the two alternating configurations able to perform their distinct set of functions and how do they compare to homodimeric kinesin-14’s such as Ncd? Here we tried to answer these questions by investigating in detail the microtubule-binding properties of Kar3Cik1 in the presence of ADP, AMPPNP, and the absence of nucleotide. Our data presented here are discussed with respect to recent structural studies on Kar3Vik1 (Rank et al., 2012; Cope et al. in press) as well as to previous EM data on homodimeric Ncd (Sosa et al., 1997; Wendt et al., 2002; Endres et al., 2006) a kinesin-14 from Drosophila melanogaster (McDonald et al., 1990; Walker et al., 1990).
Excess ADP significantly lowered the microtubule-binding affinity of Kar3Cik1 in our experiments, which is reminiscent of both Kar3Vik1 (Rank et al., 2012; Cope et al., in press) and dimeric Ncd (Sosa et al., 1997; Wendt et al., 2002). However, microtubule binding increased greatly in the absence of nucleotides or in the presence of AMPPNP. At high microtubule-affinity conditions the overall structural configurations of the three kinesin-14’s domains are very similar (see Figs. 6 & 7, and for Ncd: Hirose et al., 1996; Sosa et al., 1997; Wendt et al., 2002; Endres et al., 2006), suggesting a common mechanism for the kinesin-14 powerstroke within this kinesin family. Homodimeric Ncd has been shown to bind with only one of its motor domains to the microtubule surface while the second one protrudes outwards. This matches the domain configuration of the heterodimeric complexes where the Kar3 motor domain maintains contact to the microtubule surface while the Cik1 (wireframe in Fig. 7) or Vik1 motor homology domains (solid volume in Fig. 7) extend away from the microtubule. In the case of the Ncd homodimer, it has been argued that having two identical motor domains increases the chance of finding a microtubule binding site and that prior to binding both motors may have the same probability to become the microtubule-binding domain (Wendt et al., 2002; Liu et al., 2012). On the other hand, Foster et al. (2001) reported about asymmetry in dimeric Ncd in solution where one head held ADP tightly and the other weakly, which would enhance its probability for microtubule binding over the other one. In any case, upon binding, the motor adopts an asymmetric configuration where the microtubule-bound domain diverts its task to regular motor function, and the tethered domain adopts a helper function perhaps aiding the stalk rotation. Kar3Cik1 and Kar3Vik1, on the other hand, do not have the advantage of a dimeric motor domain, but instead maintain a distinct variability in their function by selectively forming heterodimers depending on the circumstances (reviewed in: Winey & Bloom, 2012). The initial process of finding a microtubule-binding site appears to be aided by the motor-homology domains (Allingham et al., 2007; Duan et al., 2012; Chen et al., 2012), but during the powerstroke Kar3 takes over and remains bound to the microtubule while Cik1 or Vik1 rotate together with the motor’s coiled-coil stalk in response to nucleotide uptake into Kar3’s active site.
Despite their strong structural similarities upon microtubule binding (see Figs. 6 & 7), there are small but distinct differences between the 3-D maps of microtubule-bound Kar3Cik1 and Kar3Vik1 for both the nucleotide-free and ATP binding states (see Fig. 6; circles and arrows). Most of these differences are between the tethered Cik1 (Fig. 7A & C) and Vik1 domains (Fig. 7B & D) while the tubulin and Kar3 domains are essentially identical (Figs. 6 & 7). Given their distinct functional capabilities, this result suggests that subtle structural differences between the two complexes, such as the increased flexibility seen in Kar3Cik1 over Kar3Vik1 (Fig. 6) may still result in crucial differences in motor function. Kinesin heads of various families show very little variations between each other. However, ATP binding sometimes induces a slight rotation of the bound kinesin head with respect to the tubulin protofilament (Kikkawa et al., 2001; Skiniotis et al., 2003; Hirose et al., 2006). Therefore, the variations observed between Cik1 and Vik1 may be indicative of their different cellular tasks.
Yet another notable difference between Kar3Cik1 and Kar3Vik1 are their microtubule decoration properties in both high-affinity states (AMPPNP and absence of nucleotides). Kar3Vik1 shows a very distinct, highly cooperative binding mechanism, very similar to Ncd (see Fig. 2 in Wendt et al., 2002). At sub-stoichiometric incubation conditions (Fig. 5) Kar3Vik1 completely fills individual protofilaments but may leave most of the microtubule surface free (Fig. 5A). Kar3Cik1 complexes, however, bind in a more stochastic fashion, randomly filling the microtubule surface but leaving numerous gaps in-between (Fig. 5B). The data shown in Figure 5 have been acquired in the presence of AMPPNP but the differences between Kar3Cik1 and Kar3Vik1 with regard to cooperative binding are also prevalent in the absence of nucleotide. Hence, though small, structural and functional differences are apparent and are indicative for different tasks: Kar3Cik1 promotes microtubule-depolymerization (Page & Snyder 1992; Sproul et al., 2005), while Kar3Vik1, like Ncd, maintains the mitotic spindle and adds stability to the system, possibly achieved by the cooperative binding property (Manning et al., 1999; reviewed in: Winey & Bloom, 2012). Unlike with other microtubule-depolymerizing kinesins we have never observed any type of kinesin-tubulin protofilament curls as previously found with kinesin-13 (see Moores et al., 2006; Mulder et al., 2009). Curls neither appeared with Kar3Cik1, Kar3Vik1 nor Ncd, confirming earlier data that the depolymerizing mechanism of Kar3Cik1 must be different from kinesin-13 (see Sproul et al., 2005).
Conclusions
While we have shown that the 3-D structural configurations of microtubule binding and minus-end directed movement are very similar between all three kinesin-14s compared here (and very different to other kinesins), the key to the specific cellular functions of Kar3Cik1 versus Kar3Vik1 and Ncd, may be influenced by slight conformational variations, and a difference in cooperative binding. Strong cooperative binding may be disruptive to Kar3Cik1’s function as a microtubule-depolymerizing factor, while the enhanced cooperativity observed with Kar3Vik1 and Ncd could benefit their modes of action by rapidly recruiting multiple motors to a particular site of interest, thereby facilitating spindle stabilization and compensating for the inherent lack of processivity, and overcome a situation where an isolated motor complex would be rather ineffective.
Neither this work nor the one on Kar3Vik1 (Cope et al., in press) revealed any cryo-EM 3-D data that showed stoichiometric binding of monomeric Vik1 or Cik1 constructs (not in complex with Kar3) to the microtubule at any nucleotide condition, suggesting no to little microtubule binding affinity of these domains. Furthermore, only the shadowed images of kinesin-14’s in the presence of ADP indicate a cross-protofilament binding configuration of Kar3Cik1, Ncd (Fig. 3A, B) and Kar3Vik1 (Rank et al., 2012). These results are in stark contrast to the microtubule co-sedimentation studies that show that microtubule binding by either Cik1 or Vik1 is quite strong in the presence of ADP based on the assumption that the Kar3•ADP head interacts weakly with the microtubule (Allingham et al., 2007; Duan et al., 2012; Chen et al., 2012). The reason for this discrepancy most certainly lies in the different experimental designs. which may favor capture of transient intermediates by solution equilibrium binding studies yet not by cryo-EM. The same discrepancy between EM imaging and kinetic measurements has also been found for the ADP-Ncd microtubule complex, despite the presence of two regular motor domains in this homodimer (Foster et al., 2001; Liu et al., 2012).
These results suggest that the initial microtubule collision by Cik1 or Vik1 under some conditions is transient and is followed immediately by Kar3•ADP microtubule association resulting in ADP release and destabilization of the Cik1 or Vik1 microtubule interaction (Allingham et al., 2007; Rank et al. 2012; Duan et al., 2012; Chen et al., 2012). This model explains why Cik1 and Vik1, while showing high microtubule binding affinity by them do not stall the motor. These observations emphasize the need for allosteric interactions between the motor domain and Cik1 or Vik1 to overcome their lack of an ATP site that would otherwise regulate the microtubule binding affinity and on-off rate as seen in regular kinesin motor domains.
Material & Methods
Microtubule polymerization
Microtubules were polymerized under conditions that favor the formation of 15-protofilament microtubules as described (Beuron & Hoenger, 2001). Briefly: Polymerized microtubules were made in vitro using 45 μM bovine brain tubulin (Cytoskeleton, Inc., Denver, CO), BRB80 (80 mM PIPES, pH 6.8, 1 mM MgCl2, 1 mM EGTA), 1 mM GTP, 10 μM paclitaxel (Sigma, St. Louis, MO) and 15% (v/v) DMSO that has been found to enhance the fraction of 15-protofilament microtubules. They were incubated at 37°C for 30 min and stored at room temperature overnight which we found to reduce open tubulin sheets and increase the amount of stable closed tubes.
Expression and purification of WT GCN4-Kar3Cik1
GCN4-Kar3Cik1 was expressed and purified similarly to previously reported methods (Sproul et al., 2005, Allingham et al., 2007). GCN4-Kar3Cik1 is a truncation of the full-length Kar3Cik1 containing residues Lys353 - Lys729 of Kar3, and Asn344 – Asp594 of Cik1. This construct comprises of the complete C-terminal globular domains of Kar3 and Cik1 plus part of native coiled-coil stalk through which Kar3 and Cik1 heterodimerize. Furthermore, to initialize the correct hetero-dimerization between Kar3 and Cik1 a GCN4 leucine zipper sequence (O’Shea et al., 1991) was added to the N-terminus of the truncated Kar3Cik1 construct, as recently described for Kar3Vik1 (Rank et al., 2012).
Vitrification of WT GCN4-Kar3Cik1-MT complexes for cryo-EM
Vitrification of protein complexes and data recording by cryo-EM warrants the best possible structure preservation (e.g. see Dubochet, 2007). Cryo-EM GCN4-Kar3Cik1-MT complexes were assembled directly on holey carbon C-flat grids (Protochips, Inc., Raleigh, NC). Polymerized MTs were diluted to 2.25 μM with BRB80. 5 μl of diluted MTs were allowed to adsorb to a holey carbon grid for 30s and excess liquid was blotted away. 13.55 μM GCN4-Kar3Cik1 in ATPase buffer (for nucleotide-free conditions: 20 mM HEPES pH 7.2, 5 mM magnesium acetate, 50 mM potassium acetate, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT; for AMPPNP conditions; BRB80) was added to the MTs for approximately 2 minutes, blotted with a Whatman #1 filter paper and subsequently plunge frozen in liquid ethane at liquid-nitrogen temperature (Dubochet et al., 1988).
Unidirectional metal shadowing of Kar3Cik1-MT complexes
Kar3Cik1 at 4.5 μM in ATPase buffer and 1 mM ADP was added to 3.75 μM MTs and incubated for 90–120 s. Kar3Cik1-MT complexes were adsorbed to regular 200-mesh copper EM-grids, coated with a single carbon-film and vitrified by plunging the grids into liquid ethane. Frozen grids were either stored under liquid nitrogen or subsequently transferred into the so-called Midilab freeze-drying / shadowing unit, freeze-dried for approximately 2 hours at −90C and a pressure of ~10−7 bar. Then the grids were unidirectionally shadowed with a ~0.3nm thick layer of tantalum/tungsten, transferred under cryo-vacuum conditions directly into the microscope and mounted onto a modified Gatan-626 cryo-holder (Hoenger et al., 2000).
ADP state
GCN4-Kar3Cik1 at a final concentration of 13.55 μM in ATPase buffer, 5% sucrose and 1 mM ADP was incubated at room temperature for 10 min. MTs at a final concentration of 2.25 μM were added to the GCN4-Kar3Cik1-ADP solution and incubated for another 15 mins. GCN4-Kar3Cik1-ADP - MT complexes were applied to a holey carbon grid and vitrified as described above. ADP motor-MT complexes were also prepared for shadowing experiments, described above.
Nucleotide-free state
Incubation of motors with the ATP/ADP hydrolyzing enzyme apyrase (Sigma, St. Louis, MO) generated the nucleotide-free state. GCN4-Kar3Cik1 was diluted to 13.55 μM with ATPase buffer and 1 unit of apyrase. The kinesin-apyrase mixture was incubated on ice for 30–45 min and vitrified for cryo-EM imaging with MT on holey carbon grids as described above.
ATP state
To generate a stable ATP state, ATP hydrolysis in GCN4-Kar3Cik1-MT complexes had to be prevented. This is routinely achieved by substitution of ATP with the non-hydrolyzable ATP analog adenylyl imidodiphosphate tetralithium salt (AMPPNP: see Shimizu et al., 1993) (Sigma, St. Louis, MO). GCN4-Kar3Cik1 at a final concentration 13.55 μM in ATPase buffer was incubated with 2.2 mM AMPPNP on ice for 20–30 min and frozen with MT on holey carbon grids as described above.
Cryo-EM data collection and data processing
Vitrified samples were transferred to a Gatan-626 cryo-holder (Gatan, Inc, Pleasanton, CA). Cryo-EM data was collected on an FEI Tecnai F20 FEG transmission EM (FEI-Company, Hillsboro, OR, and Eindhoven, The Netherlands) operating at 200 kV. Single frame images were collected at a nominal magnification of 29,000× and a defocus value of −2.5 μm with an electron dose of 20 e−/Å2. Images were recorded without binning on a 4K × 4K Gatan Ultrascan 895 CCD camera (Gatan, Inc, Pleasanton, CA) which, at 29,000× created a pixel size of 3.8Å with respect to the specimen. Due to the nature of helical 3-D reconstruction tilting the specimen was not necessary. Images were screened for 15-protofilament MTs and processed with helical 3-D reconstruction using the software packages PHOELIX (Whittaker et al., 1995) and SUPRIM (Schroeter and Bretaudiere, 1996). For the docking attempts shown here we were using Chimera (Pettersen et al., 2004), and we docked the tubulin high-resolution structure according to Nogales et al. (1999).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allingham JS, Sproul LR, Rayment I, Gilbert SP. Vik1 modulates microtubule-Kar3 interactions through a motor domain that lacks an active site. Cell. 2007;128:1161–1172. doi: 10.1016/j.cell.2006.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnal I, Metoz F, DeBonis S, Wade RH. Three-dimensional structure of functional motor proteins on microtubules. Curr Biol. 1996;6:1265–70. doi: 10.1016/s0960-9822(02)70712-4. [DOI] [PubMed] [Google Scholar]
- Beuron F, Hoenger A. Structural analysis of the microtubule-kinesin complex by cryo-electron microscopy. Methods Mol Biol. 2001;164:235–54. doi: 10.1385/1-59259-069-1:235. [DOI] [PubMed] [Google Scholar]
- Case RB, Pierce DW, Hom-Booher N, Hart CL, Vale RD. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell. 1997;90:959–966. doi: 10.1016/s0092-8674(00)80360-8. [DOI] [PubMed] [Google Scholar]
- Chen CJ, Rayment I, Gilbert SP. Kinesin Kar3Cik1 ATPase pathway for microtubule cross-linking. J Biol Chem. 2011;286:29261–72. doi: 10.1074/jbc.M111.255554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Rayment I, Gilbert SP. The ATPase pathway that drives the Kinesin-14 Kar3Vik1 powerstroke. J Biol Chem. 2012;287:36673–82. doi: 10.1074/jbc.M112.395590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope J, Gilbert SP, Rayment I, Mastronarde D, Hoenger A. Cryo-electron tomography of microtubule-kinesin motor complexes. J Struct Biol. 2010;170:257–265. doi: 10.1016/j.jsb.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope J, Rank KC, Gilbert S, Rayment I, Hoenger A. Kar3Vik1 Uses a Minus-End Directed Powerstroke for Movement Along Microtubules. PLOS-one. 2012 doi: 10.1371/journal.pone.0053792. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro MJ, Fondecave RM, Clarke LA, Schmidt CF, Stewart RJ. Working strokes by single molecules of the kinesin-related microtubule motor ncd. Nature Cell Biology. 2000;2:724–729. doi: 10.1038/35036357. [DOI] [PubMed] [Google Scholar]
- Duan D, Jia Z, Joshi M, Brunton J, Chan M, et al. Neck Rotation and Neck Mimic Docking in the Noncatalytic Kar3-associated Protein Vik1. J Biol Chem. 2012;287:40292–301. doi: 10.1074/jbc.M112.416529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, et al. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Dubochet J. The physics of rapid cooling and its implications for cryoimmobilization of cells. Methods Cell Biol. 2007;79:7–21. doi: 10.1016/S0091-679X(06)79001-X. [DOI] [PubMed] [Google Scholar]
- Endow SA, Henikoff S, Soler-Niedziela L. Mediation of meiotic and early mitotic chromosome segregation in Drosophila by a protein related to kinesin. Nature. 1990;345:81–83. doi: 10.1038/345081a0. [DOI] [PubMed] [Google Scholar]
- Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, et al. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 1994;13:2708–13. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres NF, Yoshioka C, Milligan RA, Vale RD. A lever-arm rotation drives motility of the minus-end-directed kinesin Ncd. Nature. 2006;439:875–878. doi: 10.1038/nature04320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G, Hajdo L, Skowronek KJ, Reuther C, Kasprzak AA, et al. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nature Cell biology. 2009;11:717–723. doi: 10.1038/ncb1877. [DOI] [PubMed] [Google Scholar]
- Foster KA, Gilbert SP. Kinetic studies of dimeric Ncd: evidence that Ncd is not processive. Biochemistry. 2000;39:1784–1791. doi: 10.1021/bi991500b. [DOI] [PubMed] [Google Scholar]
- Foster KA, Mackey AT, Gilbert SP. A mechanistic model for Ncd directionality. J Biol Chem. 2001;276:19259–66. doi: 10.1074/jbc.M008347200. [DOI] [PubMed] [Google Scholar]
- Gardner MK, Haase J, Mythreye K, Molk JN, Anderson M, et al. The microtubule-based motor Kar3 and plus end-binding protein Bim1 provide structural support for the anaphase spindle. J Cell Biol. 2008;180:91–100. doi: 10.1083/jcb.200710164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick AM, Song H, Endow SA, Rayment I. X-ray crystal structure of the yeast Kar3 motor domain complexed with Mg.ADP to 2.3 A resolution. Biochemistry. 1998;37:1769–1776. doi: 10.1021/bi972504o. [DOI] [PubMed] [Google Scholar]
- Hirose K, Lockhart A, Cross RA, Amos LA. Three-dimensional cryoelectron microscopy of dimeric kinesin and ncd motor domains on microtubules. Proc Natl Acad Sci U S A. 1996;93:9539–44. doi: 10.1073/pnas.93.18.9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K, Akimaru E, Akiba T, Endow SA, Amos LA. Large conformational changes in a kinesin motor catalyzed by interaction with microtubules. Mol Cell. 2006;23:913–923. doi: 10.1016/j.molcel.2006.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenger A, Sablin EP, Vale RD, Fletterick RJ, Milligan RA. Three dimensional Structure of a Tubulin-Motor Protein Complex. Nature. 1995;376:271–274. doi: 10.1038/376271a0. [DOI] [PubMed] [Google Scholar]
- Hoenger A, Doerhoefer M, Woehlke G, Tittmann P, Gross H, et al. Surface Topography of Microtubule Walls Decorated with Monomeric and Dimeric Kinesin Constructs. Biol Chem. 2000;381:1001–11. doi: 10.1515/BC.2000.123. [DOI] [PubMed] [Google Scholar]
- Hoenger A, Gross H. Structural investigations into microtubule-MAP complexes. Methods Cell Biol. 2008;84:425–44. doi: 10.1016/S0091-679X(07)84014-3. [DOI] [PubMed] [Google Scholar]
- Karabay A, Walker RA. Identification of microtubule binding sites in the Ncd tail domain. Biochemistry. 1999;38:1838–1849. doi: 10.1021/bi981850i. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kikkawa M, Sablin EP, Okada Y, Yajima H, Fletterick RJ, et al. Switch-based mechanism of kinesin motors. Nature. 2001;411:439–45. doi: 10.1038/35078000. [DOI] [PubMed] [Google Scholar]
- Krzysiak TC, Wendt T, Sproul LR, Tittmann P, Gross H, et al. A structural model for monastrol inhibition of dimeric kinesin Eg5. EMBO J. 2006;25:2263–73. doi: 10.1038/sj.emboj.7601108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313:1045–57. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- Maddox PS, Stemple JK, Satterwhite L, Salmon ED, Bloom K. The minus end-directed motor Kar3 is required for coupling dynamic microtubule plus ends to the cortical shmoo tip in budding yeast. Curr Biol. 2003;13:1423–8. doi: 10.1016/s0960-9822(03)00547-5. [DOI] [PubMed] [Google Scholar]
- Manning BD, Barrett JG, Wallace JA, Granok H, Snyder M. Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J Cell Biol. 1999;144:1219–1233. doi: 10.1083/jcb.144.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald HB, Goldstein LS. Identification and characterization of a gene encoding a kinesin-like protein in Drosophila. Cell. 1990;61:991–1000. doi: 10.1016/0092-8674(90)90064-l. [DOI] [PubMed] [Google Scholar]
- McDonald HB, Stewart RJ, Goldstein LS. The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell. 1990;63:1159–1165. doi: 10.1016/0092-8674(90)90412-8. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Milligan RA, Flicker PF. Structural relationships of actin, myosin, and tropomyosin revealed by cryo-electron microscopy. J Cell Biol. 1987;105:29–39. doi: 10.1083/jcb.105.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores CA, Cooper J, Wagenbach M, Ovechkina Y, Wordeman L, et al. The role of the kinesin-13 neck in microtubule depolymerization. Cell Cycle. 2006;5:1812–5. doi: 10.4161/cc.5.16.3134. [DOI] [PubMed] [Google Scholar]
- Mountain V, Simerly C, Howard L, Ando A, Schatten G, et al. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol. 1999;147:351–66. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder AM, Glavis-Bloom A, Moores CA, Wagenbach M, Carragher B, et al. A new model for binding of kinesin 13 to curved microtubule protofilaments. J Cell Biol. 2009;185:51–7. doi: 10.1083/jcb.200812052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999 Jan;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- Oladipo A, Cowan A, Rodionov V. Microtubule motor Ncd induces sliding of microtubules in vivo. Mol Biol Cell. 2007;18:3601–6. doi: 10.1091/mbc.E06-12-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–44. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- Page BD, Snyder M. CIK1: a developmentally regulated spindle pole body-associated protein important for microtubule functions in Saccharomyces cerevisiae. Genes Dev. 1992;6:1414–29. doi: 10.1101/gad.6.8.1414. [DOI] [PubMed] [Google Scholar]
- Page BD, Satterwhite LL, Rose MD, Snyder M. Localization of the Kar3 kinesin heavy chain-related protein requires the Cik1 interacting protein. J Cell Biol. 1994;124:507–19. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman EJ, Scholey JM. Mitotic microtubule crosslinkers: insights from mechanistic studies. Curr Biol. 2009;19:R1089–94. doi: 10.1016/j.cub.2009.10.047. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rank KC, Chen JC, Cope J, Porche K, Hoenger A, et al. Kar3Vik1, a member of the Kinesin-14 superfamily, shows a novel kinesin microtubule binding patter. J Cell Biol. 2012;197:957–970. doi: 10.1083/jcb.201201132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD. Nuclear fusion in the yeast Saccharomyces cerevisiae. Annu Rev Cell Dev Biol. 1996;12:663–95. doi: 10.1146/annurev.cellbio.12.1.663. [DOI] [PubMed] [Google Scholar]
- Schroeter JP, Bretaudiere JP. SUPRIM: easily modified image processing software. J Struct Biol. 1996;116:131–137. doi: 10.1006/jsbi.1996.0021. [DOI] [PubMed] [Google Scholar]
- Skiniotis G, Surrey T, Altmann S, Gross H, Song YH, et al. Nucleotide-induced conformations in the neck region of dimeric kinesin. EMBO J. 2003;22:1518–1528. doi: 10.1093/emboj/cdg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablin EP, Case RB, Dai SC, Hart CL, Ruby A, et al. Direction determination in the minus-end-directed kinesin motor ncd. Nature. 1998;395:813–6. doi: 10.1038/27463. [DOI] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-Related Proteins Required for Structural Integrity of the Mitotic Spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, et al. The Bipolar Kinesin, KLP61F, Cross-links Microtubules within Interpolar Microtubule Bundles of Drosophila Embryonic Mitotic Spindles. J Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Toyoshima YY, Vale RD. Use of ATP analogs in motor assays. Methods Cell Biol. 1993;39:167–77. doi: 10.1016/s0091-679x(08)60169-7. [DOI] [PubMed] [Google Scholar]
- Sosa H, Dias DP, Hoenger A, Whittaker M, Wilson-Kubalek, et al. A model for the microtubule-Ncd motor protein complex obtained by cryo-electron microscopy and image analysis. Cell. 1997;90:217–224. doi: 10.1016/s0092-8674(00)80330-x. [DOI] [PubMed] [Google Scholar]
- Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP. Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr Biol. 2005;15:1420–1427. doi: 10.1016/j.cub.2005.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Milligan RA. The way things move: Looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- Walker RA, Salmon ED, Endow SA. The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature. 1990;347:780–782. doi: 10.1038/347780a0. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Verma S, Mitchison TJ. XCTK2: a kinesin-related protein that promotes mitotic spindle assembly in Xenopus laevis egg extracts. J Cell Biol. 1997;136:859–70. doi: 10.1083/jcb.136.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt TG, Volkmann N, Goldie KN, Müller J, Mandelkow E, et al. Microtubule Binding Patterns of the Reverse Kinesin Motor Ncd reveal a Minus-End Directed Power Stroke. EMBO J. 2002;21:5969–5978. doi: 10.1093/emboj/cdf622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T, Karabay A, Walker RA, Gross H, Hoenger A. A Structural Analysis of the Interaction between Ncd Tail and Tubulin Protofilaments. J Mol Biol. 2003;333:541–552. doi: 10.1016/j.jmb.2003.08.051. [DOI] [PubMed] [Google Scholar]
- Whittaker M, Carragher BO, Milligan RA. PHOELIX: a package for semi-automated helical reconstruction. Ultramicroscopy. 1995;58:245–259. doi: 10.1016/0304-3991(95)00057-8. [DOI] [PubMed] [Google Scholar]
- Winey M, Bloom K. Mitotic spindle form and function. Genetics. 2012;190:1197–224. doi: 10.1534/genetics.111.128710. [DOI] [PMC free article] [PubMed] [Google Scholar]