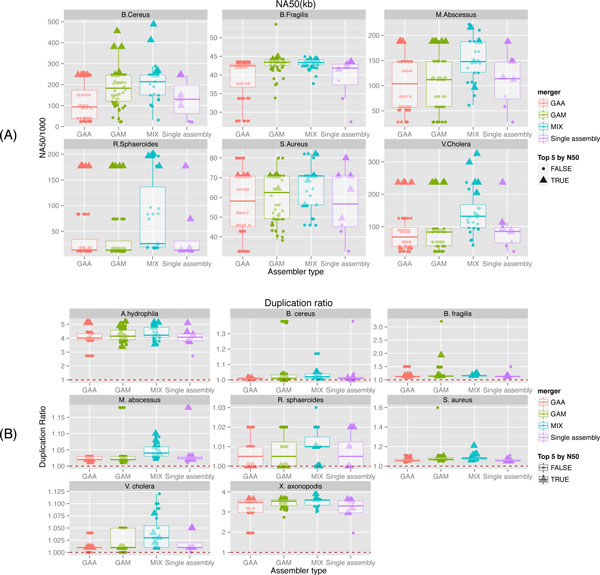
Figure 3.
Comparison of (A) NA50 and (B) duplication ratio measures for GAGE-B benchmark. (A) For six bacterial genome (six panels), eight assemblies were provided by GAGE-B, and were merged either with GAA (64 combinations), GAM-NGS (64 combinations) or Mix (28 combinations only since no asymmetry between input assemblies is introduced) or not further processed (Single Assembly). The resulting assemblies were accessed against the reference genome by QUAST and the length of the shortest aligned contig from all that cover 50% of all assembly (AKA NA50 or "corrected N50") for each possible combinations of species, mergers and assembers are reported as points (Top panel). The higher the better. Box-plots indicate the quartiles of the distribution of NA50. For each species and mergers, the top 5 combinations of assemblies according to N50 were selected, and their NA50 are depicted using large triangles. Panel (B)) report the duplication ratio of the same assemblies, the horizontal dashed line indicate a perfect ratio of 1.