Figure 3.
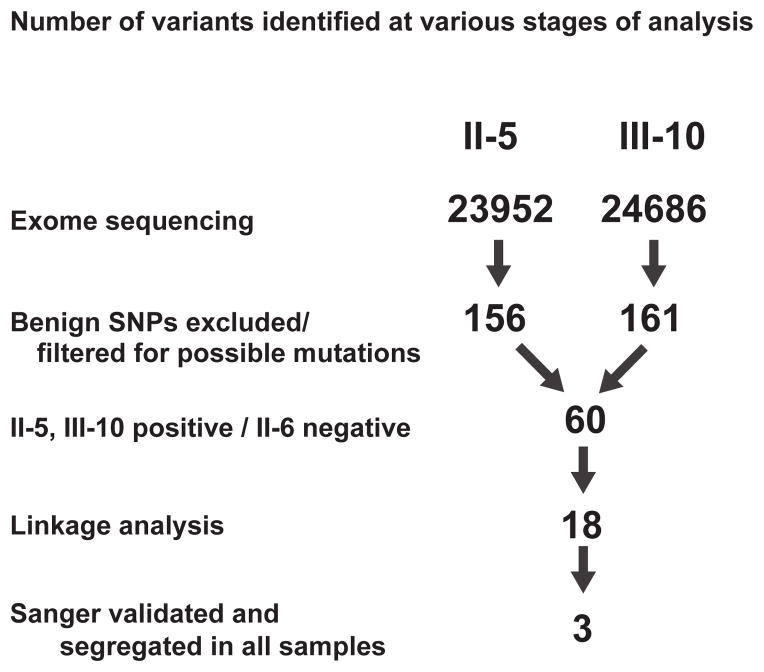
Exome sequencing revealed 23,952, 26,466, and 24,686 variants in individuals II-5 (affected), II-6 (unaffected) and III-10 (affected), respectively. After excluding reported SNPs in public databases (dbSNP 135 (http://www.ncbi.nlm.nih.gov/projects/SNP/), NHLBI Ground Opportunity Exome Sequencing Project (https://esp.gs.washington.edu/drupal/) and the 1000 Genomes Project (http://www.1000genomes.org/) and filtering for nonsynonymous, nonsense, or splice variants, or coding insertion/deletions, the numbers of candidate variants were reduced to 156, 183, and 161 in subjects II-5, II-6 and III-10, respectively. Among 60 variants that co-segregated with the phenotype of these three members of the family, there were 18 variants that were within the linkage peaks. Sanger sequencing of those 18 variants in II-5, II-6, and III-10, as well as in other family members (II-1, II-2, III-8 and III-11) confirmed the whole exome sequencing results and demonstrated that 3 of the variants co-segregated with the phenotype in the broader family (Supplemental Table 1).