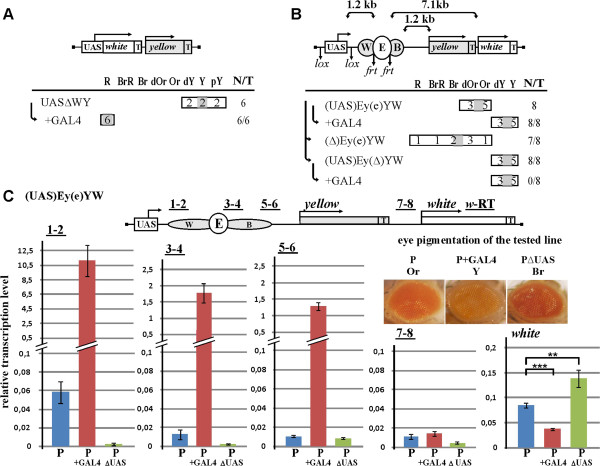
Figure 1.
The upstream UAS promoter suppresses activity of the white enhancer. (A) UAS∆WY transgenic lines. The UAS promoter drives expression of the white gene; yellow gene was used as marker to select transgenes. “T” on the 3’-side of genes indicates terminators of transcription. Below the maps, phenotypes of parental lines and those after induction of GAL4 expression (+GAL4) are shown. The color scale for white is indicated above the horizontal line. Only the range of grades that were actually recorded in the flies is shown. Each entry in the frame is the number of transgenic lines with the corresponding pigmentation grade; the shaded region in each frame indicates the “mean level.”T is the total number of lines examined; for derivative constructs, N is the number of lines where the phenotype changed as compared with the parental construct. (B) (UAS)Ey(e)YW lines. The UAS promoter drives transcription through the eye enhancer (E) of the white gene, placed between wing (W) and body (B) enhancers of the yellow gene. Downward arrows indicate lox and frt sites. Below the maps are the expression data for the parental construct and for those derived after in vivo excision of the elements. (C) Quantification of (UAS)Ey(e)YW transcripts by RT-qPCR. Positions of primer pairs (1-2, 3-4, 5-6, 7-8) are indicated. Individual transcript levels were normalized relative to ras64B for the amount of input cDNA. The transgenic material of pupae was obtained from crosses between (P) homozygous parental line and yw1118 line, (P + GAL4) homozygous parental line and GAL4-expressing line, or (P∆UAS) homozygous derivative line with deleted UAS promoter and yw1118 line. Error bars indicate standard deviations. Statistical significance was analyzed using the Student’s t-test and expressed as a P-value. **P< 0.01; ***P< 0.005. Photographs show eye pigmentation in the heterozygous parental line and its derivatives used in RT-qPCRs.