Abstract
Aims: Human γ-glutamyltranspeptidase 1 (hGGT1) is a cell-surface enzyme that is a regulator of redox adaptation and drug resistance due to its glutathionase activity. The human GGT2 gene encodes a protein that is 94% identical to the amino-acid sequence of hGGT1. Transcriptional profiling analyses in a series of recent publications have implicated the hGGT2 enzyme as a modulator of disease processes. However, hGGT2 has never been shown to encode a protein with enzymatic activity. The aim of this study was to express the protein encoded by hGGT2 and each of its known variants and to assess their stability, cellular localization, and enzymatic activity. Results: We discovered that the proteins encoded by hGGT2 and its variants are inactive propeptides. We show that hGGT2 cDNAs are transcribed with a similar efficiency to hGGT1, and the expressed propeptides are N-glycosylated. However, they do not autocleave into heterodimers, fail to localize to the plasma membrane, and do not metabolize γ-glutamyl substrates. Substituting the coding sequence of hGGT1 to conform to alterations in a CX3C motif encoded by hGGT2 mRNAs disrupted autocleavage of the hGGT1 propeptide into a heterodimer, resulting in loss of plasma membrane localization and catalytic activity. Innovation and Conclusions: This is the first study to evaluate hGGT2 protein. The data show that hGGT2 does not encode a functional enzyme. Microarray data which have reported induction of hGGT2 mRNA should not be interpreted as induction of a protein that has a role in the metabolism of extracellular glutathione and in maintaining the redox status of the cell. Antioxid. Redox Signal. 19, 1877–1888.
Introduction
γ-glutamyltranspeptidase 1 (GGT1, EC 2.3.2.2) is a cell-surface enzyme that cleaves the γ-glutamyl bond of extracellular substrates and is conserved from bacteria to humans (Fig. 1) (17, 19, 32, 45). In humans, GGT1 (hGGT1, P19440) is expressed on the apical surfaces of glands and ducts throughout the body (14). hGGT1 cleaves the γ-glutamyl bond of extracellular glutathione (γ-Glu-Cys-Gly), glutathione-conjugates, and other γ-glutamyl compounds. Most cells are unable to take up intact glutathione. The metabolism of glutathione by GGT1 releases free glutamate and the dipeptide, cysteinyl-glycine, which is hydrolyzed to cysteine and glycine by dipeptidases. The three constituent amino acids are then transported into the cell by amino-acid transporters (17, 29). The highest level of GGT1 activity is on the apical surface of the proximal tubules in the kidney (14). GGT1 knockout mice are unable to metabolize glutathione in the glomerular filtrate, and, as a result, excrete glutathione in their urine (29). They become cysteine deficient and die by 10 weeks of age due to the cysteine deficiency. Before their death, the mice develop symptoms of severe redox stress, including cataracts and increased levels of DNA damage (8, 42). In addition to its function in normal physiology, GGT1 is induced in some cells during redox stress via NFkB- and Nrf2-mediated pathways (55). It is induced in many tumors, contributing to their resistance to alkylating agents and other classes of chemotherapeutic drugs (15, 16). GGT1 is also implicated in asthma, cardiovascular disease, and diabetes (12, 30, 44).
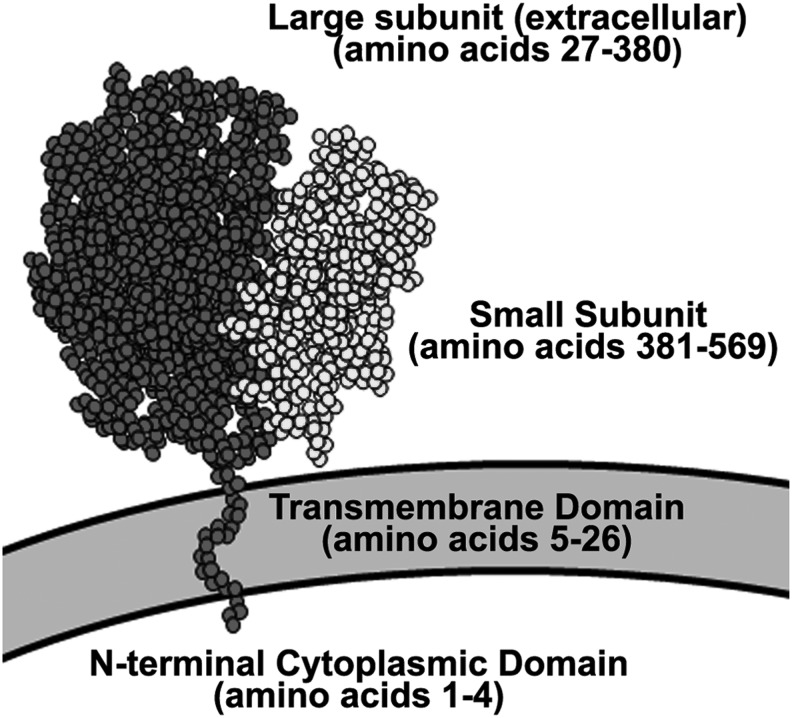
FIG. 1.
Cartoon of the structural components of mature hGGT1. hGGT1 is a type II membrane glycoprotein that undergoes autocatalytic maturation to form a heterodimeric enzyme composed of a large (a.a. 1–380, dark circles) and a small (a.a. 381–569, light circles) subunit. The transmembrane domain of the mature heterodimer is located at the N-terminus of the large subunit (a.a 5–26). The catalytic threonine residue (T381) forms the N-terminus of the small subunit. hGGT2 is predicted (www.uniprot.org/uniprot/P36268) to possess the same compositional features and structural organization as hGGT1. hGGT1, human γ-glutamyltranspeptidase 1.
Innovation.
This is the first study of the physical properties and enzymatic activity of human γ-glutamyltranspeptidase 2 (hGGT2). All three known variants of hGGT2 were expressed. Using probes specific for the large and small subunits, we demonstrated that none of the hGGT2 variants mature beyond the enzymatically inactive propeptide. We confirmed that the hGGT2 peptides are N-glycosylated, which we have shown to be required for autocleavage and maturation of hGGT1. We demonstrated that oxidative stress did not induce redox modifications that would render hGGT2 propeptides competent for auto-activation or induce a redox-sensitive chaperone or peptidase which would mediate an internal cleavage of the hGGT2 propeptide. We identified a CX3C motif that is required for optimal functional activation, and have shown that while necessary it is not sufficient to account for the lack of functional activation of the hGGT2 proteins.
The GGT2 gene is unique to humans, and it likely arises from a gene duplication event of GGT1 in recent evolutionary history (19). The initial description of a new human gene closely related to hGGT1 was first reported in 1988, but the gene was not officially named GGT2 until 2008 (18, 19, 40, 41). GGT2 bears 97% sequence identity to human GGT1 at the nucleotide level and would encode a protein (P36268) with 94% amino-acid identity to hGGT1 (19). As a result, it has been assumed that hGGT2 encodes an enzyme with the structural and functional attributes of hGGT1 (1, 19, 33, 34). This assumption recently led to the conclusion that hGGT2 is a primary regulatory enzyme which is differentially expressed to counteract oxidative stress in oncogenic K-Ras cell line models of tumorigenesis (34). hGGT2 enzymatic activity has also been proposed as a contributing factor to the potentiation of human colorectal adenocarcinoma and glaucomatous neuropathies based on pathological gene expression profiling (1, 33). However, there are no published data regarding the physical properties or enzymatic activity of the hGGT2 protein.
Similar to hGGT1, hGGT2 is predicted to be a member of the N-terminal nucleophile (Ntn) family of hydrolases and, as a result, would require an autocatalytic cleavage for its functional activation (4, 36). Characteristic of Ntn hydrolases, hGGT1 is initially expressed as a propeptide that is composed of 569 amino acids. In the early secretory pathway as the protein folds, an internal nucleophilic threonine residue (T381) initiates the autocleavage event that resolves the propeptide into two asymmetric polypeptides. These two polypeptides comprise the large (a.a. 1–380) and small (a.a. 381–569) subunits of the mature hGGT1 heterodimer (Fig. 1). After autocleavage, T381 forms the N-terminus of the small subunit and serves as the catalytic residue in the active site of hGGT1 (6). This intramolecular autocatalytic maturation is essential to the functional activation of the enzyme and is a hallmark of all known GGT orthologs, ranging from bacteria to humans (3, 6, 24, 46, 47). The mature extracellular heterodimer is held together by electrostatic interactions and remains tethered to the plasma membrane by a single-pass transmembrane domain (a.a. 5–26) that is located at the N-terminus of the large subunit (Fig. 1). The mammalian GGT1 autocleavage event is facilitated by the formation of intramolecular disulfide bonds and co-translational N-glycosylation (28, 53).
Since its initial discovery, hGGT2 mRNA sequence variants have been described (19). To determine whether hGGT2 expression leads to a functional enzyme and may contribute to the activity ascribed to hGGT1, we expressed each of the three reported cDNA variants of hGGT2 and characterized their maturation and enzymatic activity.
Results
Three mRNA coding variants for hGGT2 have been described in humans. Figure 2 shows the amino-acid sequence of hGGT1 aligned with the predicted amino-acid sequences from the three mRNA variants of hGGT2 with the ClustalW program (49). The three polypeptides encoded by these transcripts are predicted to be of disparate sizes, with theoretical molecular masses of 61.7 kDa (hGGT2-1), 62.2 kDa (hGGT2-2), and 60.7 kDa (hGGT2-3). The hGGT2-1 polypeptide is identical in length to the canonical hGGT1 propeptide, while the hGGT2-2 and hGGT2-3 variants possess insertions or deletions of 5 or 10 amino acids, respectively (Fig. 2). To determine whether the hGGT2 gene encodes an enzymatically active protein with catalytic properties similar to hGGT1, we transiently transfected the human embryonic kidney cell line, HEK293T, with cDNA constructs corresponding to each of the known hGGT2 mRNA coding variants (Fig. 2). Whole cell extracts from the transfected cells were analyzed by western blot with the GGT129 antibody, which recognizes a common epitope in the C-terminus of the large subunits of hGGT1 and all three hGGT2 variants (Fig.2). Mock-transfected HEK293T cells do not express either hGGT1 or hGGT2 or exhibit any detectable GGT activity (Fig. 3A, lane 1 and data not shown). Extracts from HEK293T cells transfected with hGGT1 cDNA contain two novel GGT129 epitopes that migrate with apparent molecular masses of 64 and 75 kDa (Fig. 3A, lane 2). We have previously shown that these bands correspond to a small population of the unprocessed hGGT1 propeptide (75 kDa) and the large subunit of mature hGGT1 (64 kDa), which exhibits a broad migration pattern due to heterogeneity in its N-glycan content (53). In contrast, each of the three hGGT2 cDNA constructs gave rise to the expression of a single, low-abundance protein band bearing the GGT129 epitope (Fig. 3A, lanes 3–5). The hGGT2-1 and hGGT2-3 proteins migrated at a position in the gel that was intermediate between the hGGT1 propeptide and mature subunit, while the hGGT2-2 protein co-migrated with the hGGT1 propeptide (Fig. 3A, lanes 2–5). These results indicate that all three hGGT2 mRNA variants encode a protein that is expressed.
FIG. 2.
Alignment of hGGT1 with the deduced amino-acid sequences of hGGT2. The amino-acid sequence of hGGT1 was aligned with the predicted amino-acid sequences from the XM_001129425.3 (hGGT2-1), XM_001129377.3 (hGGT2-2), and ENST00000405188 (hGGT2-3) mRNA variants of hGGT2. The conserved single-pass transmembrane domain is underlined. Cysteine residues (C50, C74, C192, and C196) that have been shown to form disulfide bonds in mammalian GGT1 are boxed. The conserved CX3C motif in eukaryotic orthologs of hGGT1, which has been altered in hGGT2, is shaded in gray. The seven N-glycosylation sites in hGGT1 are highlighted with a black box containing a white “N.” The catalytic threonine residue that forms the N-terminus of the small subunit (T381) after autocleavage of the hGGT1 propeptide into the mature heterodimeric enzyme is highlighted with a black box containing a white “T” with an arrow above. The epitope recognized by the GGT129 antibody is underlined with a bracket.
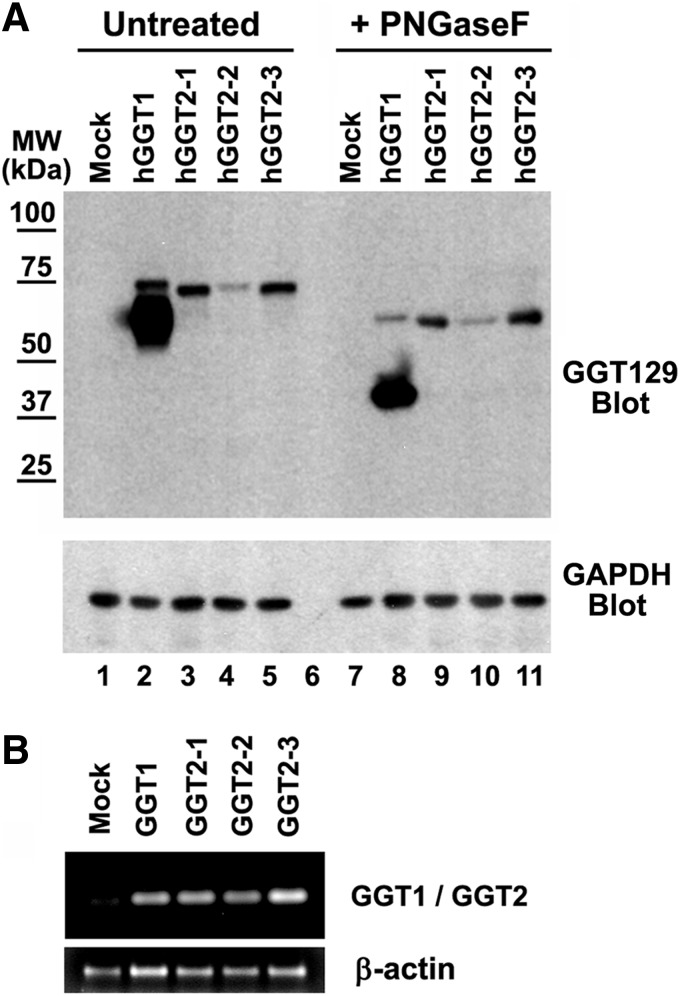
FIG. 3.
hGGT2 is expressed as an inactive propeptide that fails to autocleave. (A) Whole cell lysates from HEK293T cells transiently transfected with either empty vector (lanes 1 and 7), hGGT1 cDNA (lanes 2 and 8), hGGT2-1 cDNA (lanes 3 and 9), hGGT2-2 cDNA (lanes 4 and 10), or hGGT2-3c DNA (lanes 5 and 11) expression constructs were incubated in the presence (lanes 7–11) or absence (lanes 1–5) of PNGaseF and resolved on an 8% SDS-polyacrylamide gel. Resolved proteins were electroblotted onto nitrocellulose and then subjected to immunoblotting, using the GGT129 antibody, which recognizes a common epitope in hGGT1 and each of the hGGT2 alleles. MW, molecular-weight markers. The antibody identifies both the propeptide and large subunit of hGGT1. hGGT2-1,2,3 are present only as a propeptide. (B) Reverse transcription–polymerase chain reaction amplification of hGGT1 and hGGT2 (206 bp) or β-actin (512 bp) from transiently transfected HEK293T cells.
We then examined whether the hGGT2 proteins undergo cleavage and glycosylation. hGGT1 contains seven N-glycosylation sites (six in the large subunit and one in the small subunit), all of which are co-translationally glycosylated in HEK293 cells (53). All seven of these N-glycosylation sites are conserved in each of the three hGGT2 cDNA variants (Fig. 2). To determine whether hGGT2 is glycosylated, we incubated the extracts from the transfected HEK293T cells with the N-glycosidase, PNGaseF. After deglycosylation, the apparent molecular masses of the propeptide and large subunit of hGGT1 decreased to 61 and 42 kDa, respectively, which was consistent with the predicted molecular masses of their unglycosylated polypeptides (Fig. 3A, lane 8) (53). After incubation with PNGaseF, hGGT2-1, hGGT2-2, and hGGT2-3 also showed a decrease in apparent molecular weight, indicating that these polypeptides were also N-glycosylated (Fig. 3A, lanes 9–11). The deglycosylated hGGT2-1, hGGT2-2, and hGGT2-3 variants co-migrated with the deglycosylated hGGT1 propeptide at ∼61 kDa, which was consistent with the predicted molecular masses of the unglycosylated propeptides, 61.7, 62.2, and 60.7 kDa, respectively (Figs. 2 and 3A, lanes 9–11). No evidence for a cleaved form of any hGGT2 variant was present. The fact that they co-migrated with the hGGT1 propeptide rather than the large subunit of hGGT1 demonstrates that hGGT2 propeptides did not undergo autocleavage.
In the propeptide state, hGGT1 and other Ntn hydrolase family members are enzymatically inactive (47, 53). However, in its mature, heterodimeric form, hGGT1 can catalyze two types of biochemical reactions. Under physiological conditions, the primary reaction catalyzed by hGGT1 is the hydrolysis of compounds containing γ-glutamyl bonds, releasing glutamate from the substrate (9, 11). In the presence of high concentrations of dipeptides and some amino acids, hGGT1 can also catalyze a transpeptidation reaction, transferring the γ-glutamyl moiety to an acceptor amino acid to form a new γ-glutamyl compound (48). Extracts from HEK293T cells transfected with hGGT1 cDNA exhibited robust activity in the standardized GGT activity assays for hydrolysis and transpeptidation (specific activity of 21 mU/mg of total protein and 1.35 U/mg of total protein, respectively). However, none of the extracts prepared from HEK293T cells transfected with the three hGGT2 variants exhibited any detectable levels of either hydrolysis or transpeptidation activity. These results indicate that, despite their high sequence identity with hGGT1, including conservation of the catalytic Thr381 residue, the hGGT2 polypeptides do not catalyze GGT reactions. These data are consistent with the western blot data shown in Figure 3. Together, these data indicate that the hGGT2 propeptides do not autocleave into mature heterodimers.
Despite the disparate levels of the hGGT1 protein (present as both propeptide and mature large subunit) compared with the hGGT2 proteins (present only as propeptides), the mRNAs of all of these cDNA constructs were present at similar levels in HEK293T cells (Fig. 3B). This indicates that the differences in the protein levels of hGGT1 and the hGGT2 variants in these cells are not attributable to differences in their mRNA expression profiles.
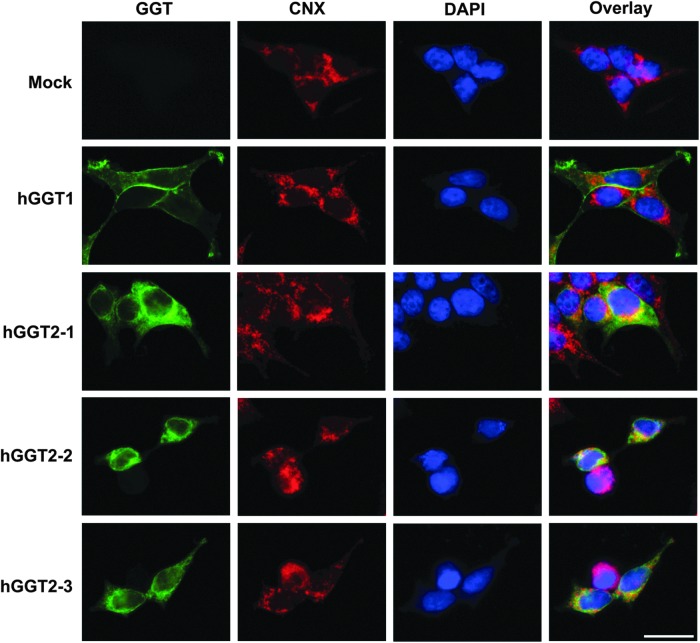
hGGT1 expressed in HEK293T cells localized to the plasma membrane (Fig. 4), which is consistent with its cellular localization pattern in normal human tissues (14). In contrast, hGGT2-1, hGGT2-2, and hGGT2-3 did not localize to the plasma membrane (Fig. 4, exposure time for GGT staining of hGGT1-transfected cells was shorter than for hGGT2 variants due to differences in the amount of protein expressed). The hGGT2 proteins accumulated in the cytoplasm in a perinuclear localization pattern that showed extensive overlap with the ER chaperone, calnexin, suggesting retention of the hGGT2 proteins in the ER (Fig. 4). This localization pattern was unexpected due to the absolute sequence conservation between the N-termini (amino acids 1–42), including the transmembrane domain, of hGGT1 and each of the hGGT2 variants (Fig. 2). These results suggest that, in the absence of autocleavage, these glycosylated polypeptides are arrested in the early secretory pathway and the hGGT2 proteins never reach the plasma membrane.
FIG. 4.
Intracellular localization of hGGT1 and hGGT2. HEK293T cells transfected with hGGT1 or hGGT2 cDNA expression constructs and grown on coverslips were fixed and processed for indirect immunofluorescence, using the GGT129 antibody (GGT, green) and an antibody against the ER marker, calnexin (CNX, red). Nuclei were stained with DAPI (blue). Areas of overlapping GGT129 and calnexin immunofluorescence appear yellow in the overlay panels. Exposure times for GGT images differ among the samples due to the lower expression levels of the hGGT2 proteins. hGGT1 is localized to the cell membrane, and hGGT2 proteins co-localize with calnexin in the ER. Scale bar=20 μm and applies to all images shown. ER, endoplasmic reticulum.
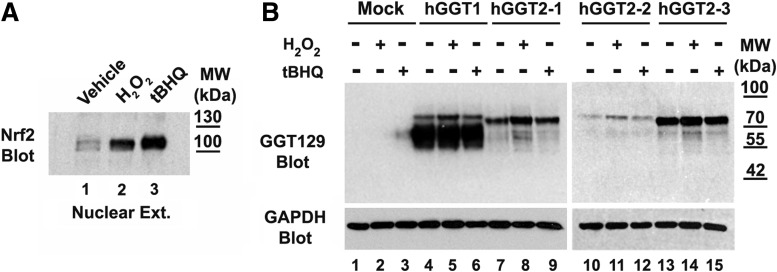
In a previous study, gene expression profiling identified hGGT2 as a potential mediator of oxidative stress resistance induced by exposure to hydrogen peroxide (H2O2) in a transformed epithelial cell line (34). We investigated whether oxidative stress induces redox modifications that would render hGGT2 propeptides competent for auto-activation or induces a redox-sensitive chaperone or peptidase which would mediate an internal cleavage of the GGT2 propeptide. Both H2O2 (250 μM) and tert-butylhydroquinone (tBHQ) (50 μM) elicited a strong oxidative stress response in HEK293T cells by 4 h of exposure, as evidenced by the stabilization and nuclear accumulation of the antioxidant response transcriptional factor, Nrf2 (Fig. 5A). We expressed each of the hGGT2 variants in the presence of either H2O2 or tBHQ for 4 and 24 h. HEK293T cells were transiently transfected with GGT1 or one of the three hGGT2 variants. Twenty-four hours after transfection, the cells were treated with 250 μM H2O2 or 50 μM tBHQ. At 4 and 24 h, cells were harvested and analyzed for processing of the GGT propeptides. The data from both time points showed that none of the hGGT2 propeptides were processed into mature hGGT2 heterodimers. The data from the 24 h time point are shown in Figure 5B. The 24 h time point was included to assess accumulation of any processed hGGT2 propeptides. Processed GGT1 is a very stable protein, with a half-life of 35 h in vivo in the rat (5). A trace amount of hGGT2-1 and hGGT2-3 appears to have been processed in the samples treated with H2O2 (Fig. 5B); however, there was no GGT catalytic activity in any of the hGGT2-1, 2-2 or 2-3 cell extracts at either 4 or 24 h. These data demonstrate that induction of oxidative stress does not alter the processing of any of the hGGT2 propeptides to yield an enzymatically active heterodimer.
FIG. 5.
Oxidative stress does not induce cleavage of hGGT2 propeptides into heterodimers. (A) Nuclear lysates (10 μg total protein) from vehicle-treated HEK293T cells (lane 1), HEK293T cells cultured for 4 h in 250 μM H2O2 (lane 2) or 50 μM tBHQ (lane 3) were subjected to SDS-PAGE and immunoblotted with an antibody specific for Nrf2. Presence of Nrf2 in the nucleus of the H2O2 and tBHQ-treated cells confirms the induction of oxidative stress under these treatment conditions. (B) HEK293T cells were transiently transfected with empty vector (lanes 1–3), hGGT1 cDNA (lanes 4–6), hGGT2-1 cDNA (lanes 7–9), hGGT2-2 cDNA (lanes 10–12), or hGGT2-3 cDNA (lanes 13–15) expression constructs. Twenty-four hours after transfection, vehicle (ethanol), 250 μM H2O2, or 50 μM tBHQ was added to the cells. After 24 h, whole cell lysates were resolved on an 8% SDS-polyacrylamide gel. Resolved proteins were electroblotted onto nitrocellulose and then subjected to immunoblotting, using the GGT129 antibody, which recognizes a common epitope in hGGT1 and each of the hGGT2 alleles. The antibody identifies both the propeptide and large subunit of hGGT1. hGGT2-1,2,3 are expressed but present only as a propeptide under each of these culture conditions. The weak signal observed in lane 3 is carried over from lane 4 as confirmed by independent blots and activity assays. H2O2, hydrogen peroxide; tBHQ, tert-butylhydroquinone.
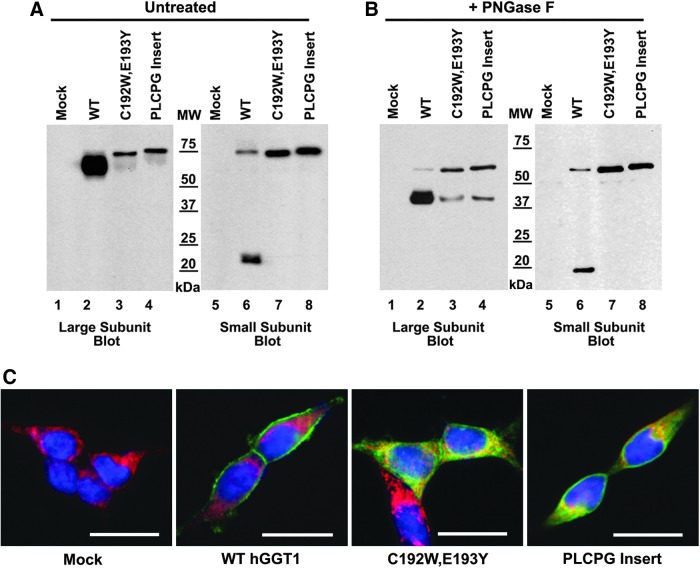
All reported transcript variants for hGGT2 possess sequence deviations in a conserved mammalian GGT1 CX3C motif (amino acids 192–196 in hGGT1; Fig. 2), which is important for the autocleavage of rat GGT1 (28). To discern whether these sequence deviations contribute to defective maturation of the hGGT2 propeptides, we employed site-directed mutagenesis to alter the amino-acid sequence in the CX3C motif in hGGT1 to match that in the hGGT2 mRNA variants. The resultant constructs encoded for hGGT1 mutants bearing either C192W plus E193Y substitutions, matching the hGGT2-1 and hGGT2-3 alleles, or a PLCPG amino-acid insertional mutation between residues C192 and E193, which mimics the hGGT2-2 primary structure at this position (Fig. 2). These mutant hGGT1 constructs were transiently transfected into HEK293T cells, and their capacity to form mature, enzymatically active hGGT1 heterodimers was evaluated. Western blot analysis with the GGT129 antibody revealed that cells transfected with wild-type hGGT1 expressed a broad protein band at 64 kDa (the mature large subunit) and a very faint band at 75 kDa (the propeptide), whereas each of the hGGT1 mutants was expressed as an hGGT1 polypeptide at a molecular weight which was consistent with the propeptide (Fig. 6A, lanes 2–4). To determine unequivocally whether the single polypeptide observed for each of the mutant constructs represented the propeptide form of the enzyme, the blots were also probed with antibodies that recognize the small subunit of hGGT1. The propeptide contains both the large and small subunits. Immunoblotting with the small subunit antibody in extracts that expressed wild-type hGGT1 protein detected two bands, the mature small subunit at 22 kDa and a low level of the hGGT1 propeptide at 75 kDa (Fig. 6, lane 6). In contrast, the extracts with the hGGT1 peptides bearing hGGT2 CX3C substitutions had only one protein (73 or 75 kDa) that was recognized by the small subunit antibody (Fig. 6A, lanes 7 and 8). The fact that these bands were recognized by both the large and small subunit antibodies demonstrates that these bands are propeptides containing both the large and the small subunits. Trace amounts of the mature large subunit were detected for the mutant hGGT1 proteins, using the large subunit antibody (apparent molecular mass of 64 kDa, Fig. 6A lanes 3 and 4), indicating a very low level of processing of the propeptide. This was also observed in the deglycosylated samples (Fig. 6B, lanes 3 and 4). The amount of the mature small subunit in these samples was too low to be detected with the small subunit antibody (Fig. 6A, lanes 7 and 8).
FIG. 6.
hGGT1 CX3C mutants do not autocleave and mislocalize to the ER. (A) Whole cell lysates from HEK293T cells transfected with either empty vector (lanes 1 and 5) or cDNA expression constructs for wild-type hGGT1 (WT, lanes 2 and 6), hGGT1(C192W;E193Y) (lanes 3 and 7), or hGGT1(C192_E193insPLCPG) (lanes 4 and 8). Five micrograms of protein from each whole cell lysate were subjected to SDS-PAGE and immunoblotting, using antibodies specific for either the large (lanes 1–4) or small (lanes 5–8) subunits of hGGT1. The introduced mutations were in the large subunit of hGGT1 and were outside the epitope recognized by the large subunit antibody. The small subunit in all constructs was identical to wild-type hGGT1 (B) Lysates from (A) were incubated with PNGaseF and subjected to SDS-PAGE and immunoblotting against either the large (lanes 1–4) or small (lanes 5–8) subunits of hGGT1. (C) HEK293T cells transfected with either empty vector, wild-type hGGT1 cDNA, or hGGT1 CX3C mutant cDNA expression constructs were grown on coverslips and fixed and processed for indirect immunofluorescence, using the GGT129 antibody (GGT, green) and an antibody against the ER marker, calnexin (CNX, red). Nuclei were stained with DAPI (blue). Areas of overlapping GGT129 and calnexin immunofluorescence appear yellow in the overlay panels shown here. Exposure times for GGT images differed among the samples due to the lower expression levels of the mutant hGGT1 proteins. hGGT1 is localized to the cell membrane, and hGGT1 CX3C mutant proteins co-localize with calnexin in the ER. Scale bars=20 μm. For the full complement of images used to compose the overlay panels, see Supplementary Figure S1.
hGGT1 is N-glycosylated. Glycosylation adds to the molecular mass of proteins to an extent that is difficult to predict. Therefore, we deglycosylated the hGGT1 proteins to assess their apparent molecular masses relative to the predicted molecular masses of the unmodified polypeptides. After deglycosylation by PNGaseF, immunoblotting with either the large or small subunit antibody showed that the dominant band from the hGGT1(C192W;E193Y) mutant migrated with an apparent molecular mass (62 kDa) identical to that observed for the deglycosylated wild-type hGGT1 propeptide (Fig. 6B, lanes, 2–3 and 6–7). These data are consistent with the molecular mass predicted from the amino-acid sequence of the unprocessed propeptide. The deglycosylated hGGT1(C192_E193insPLCPG) mutant polypeptide migrated more slowly than these two constructs (Fig. 6B, lanes 2–4 and 6–8). These results are consistent with the predicted differences in the molecular mass of the propeptide form of each of these hGGT1 proteins. Before deglycosylation, the propeptide of the hGGT1(C197W;E193Y) mutant migrated more rapidly than the wild-type allele (apparent molecular mass 73 versus 75 kDa, Fig. 6A, lanes 6–7), while the propeptide of the larger hGGT1(C192_E193insPLCPG) mutant co-migrated with the wild-type allele, with an apparent molecular mass of 75 kDa (Fig. 6A, lanes 6 and 8). These small differences in apparent mass likely reflect differences in the extent of glycosylation or the composition of the N-glycans on the mutant hGGT1 propeptides relative to the wild-type hGGT1 propeptides. These data confirm that the CX3C motif has a critical role in autocleavage and that the corresponding amino-acid sequences in the hGGT2 proteins do not fulfill this function.
Immunolocalization of the hGGT1 CX3C mutants in HEK293T cells revealed that both sets of mutations result in a failure of the hGGT1 mutant proteins to localize to the plasma membrane. The mutant GGT1 proteins accumulated in the perinuclear compartment, exhibiting considerable overlap with the ER marker, calnexin (Fig. 6C). The localization pattern for these hGGT1 CX3C mutants mirrored that observed for the wild-type hGGT2 alleles (Fig. 4). This subcellular distribution pattern suggests that perturbation of the CX3C motif and/or its effect on autocleavage prevents hGGT1 from localizing to the plasma membrane.
Biochemical analyses of HEK293T extracts transfected with the hGGT1 CX3C mutants revealed that the C192W and E193Y double mutations resulted in a 45-fold decrease in enzymatic activity relative to the wild-type allele (specific activity of 0.03 U/mg of total protein versus 1.35 U/mg of total protein, respectively), while the PLCPG insertional mutation exhibited an ∼68-fold decrease in activity (specific activity of 0.02 mU/mg of total protein). These results are consistent with the very low levels of mature hGGT1 large and small subunits detected in these extracts (Fig. 6). Taken together, these results indicate that mutating the CX3C motif of hGGT1 to match GGT2 polypeptides inhibits the maturation and proper cellular localization of hGGT1.
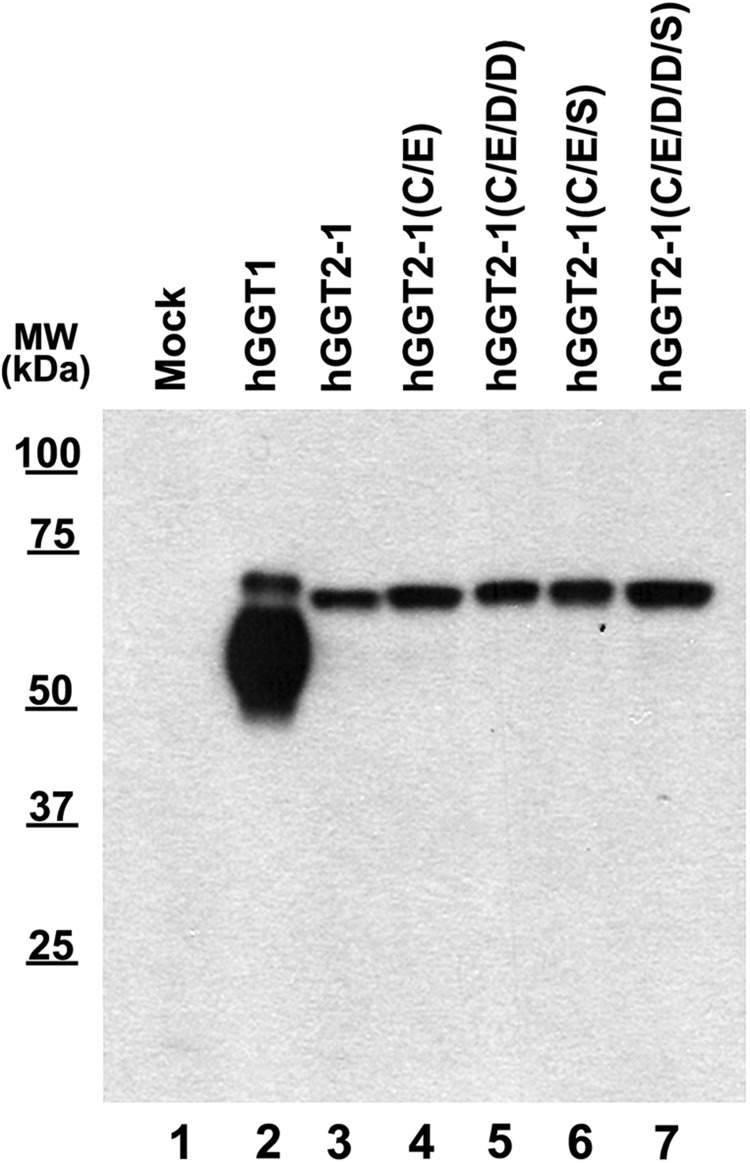
These data suggest that maintaining the sequence fidelity of the CX3C motif is critical for the proper folding and autocleavage of hGGT1. To determine whether the amino-acid sequence of the CX3C motif in hGGT1 is sufficient for conferring maturation competence to hGGT2, we reciprocally mutated the corresponding residues in the hGGT2-1 variant to the wild-type hGGT1 form, using site-directed mutagenesis. When the resulting construct, hGGT2-1(W192C;Y193E), was expressed in HEK293T cells, it failed to autocleave (Fig. 7, lane 4) or exhibit detectable levels of enzymatic activity. Similar results were observed when the hGGT2-3(W192C;Y193E) mutant was expressed in HEK293T cells (data not shown). These results demonstrate that, while the sequence fidelity of this motif is an important component of the functional maturation of hGGT1, reinstatement of the CX3C motif in hGGT2 alone is not sufficient for maturation to a heterodimer.
FIG. 7.
Targeted replacement of hGGT2 residues with wild-type hGGT1 amino acids does not restore autocatalytic maturation. Whole cell lysates from HEK293T cells transfected with either empty vector (lane1), wild-type hGGT1 cDNA (lane 2), GGT2-1 cDNA (lane 3), or mutant GGT2-1 alleles (lanes 4–7) were resolved by SDS-PAGE and electroblotted onto nitrocellulose. Immunoblotting was carried out using the GGT129 antibody. For labeling the hGGT2-1 lanes: hGGT2-1, wild-type hGGT2-1; hGGT2-1(C/E), hGGT2-1(W192C;Y193E); hGGT2-1(C/E/D/D), hGGT2-1(W192C;Y193E;T422D;T423D); hGGT2-1(C/E/S), hGGT2-1(W192C;Y193E;L451S); and hGGT2-1 (C/E/D/D/S), hGGT2-1(W192C;Y193E;T422D;T423D;L451S).
An expanded comparative analysis between the coding regions of hGGT1 with each of the hGGT2 variants identified three additional sequence deviations that are likely to impact autocleavage of hGGT2. Previous studies showed that mutagenizing either of the aspartate residues at positions 422 and 423 or the serine residue at position 451 in hGGT1 results in the expression of inactive forms of the enzyme (22, 23). All annotated mRNA variants of hGGT2 bear conserved sequence deviations at each of these three key residues, which might contribute to the inability of the hGGT2 propeptides to reach functional maturation (Fig. 2). Therefore, we introduced the corresponding amino acids from hGGT1 into the hGGT2-1(W192C;Y193E) cDNA expression construct to assess whether substituting these key residues in conjunction with the wild-type hGGT1 CX3C motif conferred functionality to hGGT2. However, when the T422D/T423D or L451S mutations were expressed either individually or in tandem within the context of the hGGT2-1(W192C;Y193E) polypeptide, the glycosylated polypeptide still failed to autocleave (Fig. 7, lanes 5–7). These mutational analyses indicate that there are multiple sequence deviations between hGGT2 and its progenitor, hGGT1, which contribute to the inability of the hGGT2 proteins to autocatalyze and become functionally mature enzymes.
Discussion
To determine whether hGGT2 encodes an enzymatically active GGT protein, we transiently expressed the known cDNA variants of hGGT2 in HEK293T cells under conditions that have previously been shown to elicit robust expression of hGGT1 (53). Reverse transcription–polymerase chain reaction (RT-PCR) analysis of the mRNA produced from each of the three hGGT2 expression constructs showed mRNA levels comparable to those observed for hGGT1 (Fig. 3B). However, unlike the hGGT1 expression construct, the translated glycoproteins expressed from the hGGT2 cDNAs failed to mature beyond the propeptide form. Data from our laboratory had previously documented that co-translational N-glycosylation is required for the autocleavage and functional maturation of the hGGT1 propeptide (53). All seven of the N-glycosylation sites from hGGT1 are conserved in all three GGT2 sequences, and we show here that the hGGT2 propeptide is also N-glycosylated (Fig. 3A). The relative mobility shift of the hGGT2 propeptides after deglycosylation was not as dramatic as that observed for the deglycosylated hGGT1 propeptide, indicating that the extent or composition of the N-glycans in the uncleaved hGGT2 propeptide may differ from that observed in hGGT1 (Figs. 2 and 3A).
We observed low levels of hGGT2 proteins relative to those observed for hGGT1 in HEK293T extracts (despite similar mRNA expression levels). We also observed aberrant intracellular localization of the expressed hGGT2 proteins. These data indicate that the glycosylated hGGT2 propeptides likely fail to fold properly and arrest in the ER where they are shepherded by resident chaperones for targeted degradation (31, 54). The only evidence that hGGT2 transcripts are actively translated in vivo is a report in which a single tryptic peptide unique to the amino-acid sequence of hGGT2 was identified during a high-throughput mass spectrometric characterization of human liver glycopeptides (7). However, we did not detect hGGT2-specific peptides within the pool of tryptic fragments from protein immunopurified from normal human kidney and liver with our GGT129 antibody (51, 52). This antibody was developed against a peptide that is identical in hGGT1 and all three variants of hGGT2 [Fig. 3A and (52)]. Our high-throughput mass spectrometric analysis of peptides and glycopeptides originating from proteins immunopurified with the GGT129 antibody did not detect any hGGT2-specific tryptic peptides despite the fact that several tryptic peptides and glycopeptides such as LAFASMFNSSEQSQK and LADTYEMLAIEGAQAFYNGSLMAQIVK are encoded by all three variants of hGGT2, but not hGGT1 (7).
Comparative sequence analyses point to several key deviations in the open reading frames of the hGGT2 propeptides that may account for the inability of the protein to reach the proper structural and/or functional status which is required for its autoactivation. Using site-directed mutagenesis, Kinlough and coworkers previously demonstrated that point mutations introduced within a conserved CX3C motif in rat GGT1 (residues 192–196 in hGGT1) resulted in the expression of a mutant hGGT1 propeptide which exhibited rate-limiting defects in its ability to autocleave into a functional enzyme (28). All known hGGT2 propeptides contain either a C192W substitution or a five-amino-acid insertional element between residues 192 and 193 that disrupt this motif (Fig. 2). In this study, when these same elements were introduced within the amino-acid sequence of hGGT1, the enzyme exhibited a defect in propeptide maturation and a dramatic decline in the detectable levels of hGGT1 protein. This demonstrates that the sequence fidelity of the CX3C motif is required for optimal functional activation (Fig. 5).
We also evaluated sequence deviations in hGGT2 that map to residues within the active site of hGGT1 and have been previously shown to be required for GGT1 enzyme function (22, 23). Using hGGT1 as a point of reference, these deviations (D422T, D423T, and S451L) map to conserved residues that have been previously shown to be required for coordinating substrate binding in the crystal structures of bacterial GGT1 orthologs (Fig. 2) (35, 37, 50). However, when these residues were replaced in hGGT2 by the corresponding residues of wild-type hGGT1, the resulting hGGT2 mutants still failed to mature into active heterodimers (Fig. 7). Therefore, it is likely that hGGT2 has acquired additional substitutions which contribute to its inability to fold properly or autocleave similar to hGGT1 and other members of the Ntn hydrolyase family (4). In addition to the sequence deviations documented earlier, hGGT2 propeptides also contain several conserved substitutions with unknown functional implications in the sequence spanning residues 421 to 435, using hGGT2-1 as the reference sequence (Fig. 2). These residues map to regions of hGGT1 that are predicted to either interact with the bound substrate in the active site or form a part of a putative “lid loop” structure which is predicted to regulate access to the catalytic nucleophile (Thr-381 in hGGT1). As a result, these sequence deviations may contribute to the failure of the hGGT2 propeptides to achieve functional maturity (21, 37).
In a recent study, Moon et al. identified induction of hGGT2 as a critical component of the enhanced antioxidant status of cells expressing the oncogenic allele of K-Ras (34). In their study, siRNA against hGGT2 enhanced H2O2-induced apoptosis of K-Ras transformed cells. However, the siRNA construct employed by Moon et al. exhibits greater than 97% complementarity to hGGT1 mRNA, suggesting that hGGT1 may have also been a target for the siRNA-mediated degradation. Data from studies of both mouse and rat cell lines show that GGT1 expression is induced by activated K-Ras (10, 39). Moon et al. suggested that induction of hGGT2 activity led to resistance to H2O2 by enhancing the antioxidant status of the cell. They did not analyze the hGGT2 protein expressed in their studies (34). Our experimental findings show that expression of hGGT2 does not result in an active enzyme. When we subjected the cells to oxidative stress with two different compounds, including the H2O2 used by Moon et al., no processing of any of the hGGT2 propeptides into enzymatically active heterodimers was observed. It is likely that the effects that Moon et al. observed in their studies were attributable to induced hGGT1 expression rather than hGGT2 expression. Expression of GGT1 has been shown to increase the antioxidant status of the cell and render cells resistant to toxins (16, 25, 42). Our data will assist in de-convoluting functional interpretations based on both published and future studies in which altered hGGT2 expression patterns are observed.
An additional source of confusion arises from inconsistencies in the nomenclature used to describe GGT-related genes (19). γ-glutamyltranspeptidase 3 pseudogene (GGT3P, NR_003267), a noncoding RNA, was at one time annotated in the NCBI database as GGT2. In 2011, Baginski et al. reported that GGT1 and GGT2 expression is co-regulated in human lung epithelium (2). However, based on their primers (targeted against Accession number NM_002058 which was discontinued and replaced by NR_003267.1), their data show mRNA levels of GGT1 and GGT3P. This same discrepancy is present in a study of p63-inducible promoters in human embryonic kidney 293 cells (38). The authors described induction of GGT2, but refer to XM_290331, an accession number that has also been discontinued and replaced by NR_003267.1, which is GGT3P. The literature is likely to continue to contain errors, because commercially available plasmids referred to as the “GGT2” plasmids (NM_002058.1) are actually GGT3P (OriGene Cat #SC303113). Similarly, commercially available vector-based GGT2 siRNAs (NM_002058.1) are directed against GGT3P (GGT2 siRNA, GenScript). To advance our understanding of human GGT genes, it is imperative that there is strict adherence to the annotative nomenclature which is used to describe them as outlined by Heisterkamp et al. in collaboration with the HUGO Gene Nomenclature Committee (19).
Our studies show that all three of the known variants of hGGT2 can be translated into proteins. However, none of these proteins have GGT activity due to a failure of the propeptide to undergo the autocatalytic cleavage needed to produce a mature, enzymatically active heterodimer. Expression of hGGT2 mRNA should not be interpreted as a measure of GGT activity within a cell or tissue.
Materials and Methods
Construction of hGGT1 and hGGT2 expression plasmids
The pcDNA 3.1(+) hGGT1 expression construct was previously described (53). The hGGT2-1 (NCBI accession no. XM_001129425.3) cDNA expression construct was synthesized by DNAExpress (Dorval, Montreal, Quebec, Canada) directly into a pcDNA3.1(+) plasmid backbone between the EcoRI and XhoI restriction sites with the same Kozak sequence (TGAGCCACC) used for the hGGT1 expression construct. The hGGT2-2 (NCBI accession no. XM_001129377.3) cDNA was synthesized by Genscript (Piscataway, NJ) and cloned into a pUC57 shuttling vector in the EcoRI restriction site. The resulting hGGT2 ORF was cloned from the pUC57 vector into pcDNA3.1 (−) expression construct between the ApaI and KpnI sites. The same Kozak sequence (TGAGCCACC) used for the hGGT1 and hGGT2-1 cDNA expression constructs was inserted immediately upstream of the coding sequence and downstream of the ApaI restriction site via the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA) with the primers listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/ars). The hGGT2-3 (Ensembl accession no.ENST00000405188) cDNA expression construct was generated by deleting the nucleotide sequence encoding amino acids 404–413 in the XM_001129425.3 cDNA ORF, using the QuikChange kit and the primer pairs listed in Supplementary Table S1. The hGGT1(C192W; E193Y) and hGGT1(C192_E193insPLCPG) mutants were generated by site-directed PCR mutagenesis, using the QuikChange site-directed mutagenesis kit with the wild-type hGGT1 cDNA expression construct as the template and the appropriate primer pairs listed in Supplementary Table S1. The hGGT2-3 (W192C;Y193E) and hGGT2-1 (W192C; Y193E) mutants were generated as described earlier, using the wild-type hGGT2-1 or hGGT2-3 cDNA expression constructs as templates, while the hGGT2-1 (W192C; Y193E; T422D; T423D), hGGT2-1 (W192C; Y193E; L451S) were generated using the hGGT2-1 (W192C; Y193E) cDNA template and the appropriate mutagenic primers (Supplementary Table S1). The hGGT2-1 (W192C; Y193E; T422D; T423D; L451S) expression construct was generated by site-directed mutagenesis, using the hGGT2-1 (W192C; Y193E; T422D; T423D) cDNA template and the L451S mutagenic primer pair (Supplementary Table S1). The sequence fidelity of each expression construct was confirmed by the DNA Sequencing Facility at the Oklahoma Medical Research Foundation (Oklahoma City, OK).
Transient expression of hGGT constructs in HEK293T cells
HEK293T cells (human embryonic kidney, ATCC # CRL-1573) were cultured in Dulbecco's Modified Eagle's Medium (DMEM) containing 10% fetal bovine serum and penicillin/streptomycin (50 units/ml, 50 μg/ml) at 37°C in the presence of 5% CO2. The cells were transfected with 10 μg of the appropriate expression plasmid, according to the calcium phosphate method (13). At 48 h, the transfectants were harvested in PBS or fixed for immunocytochemical analysis. For experiments involving oxidative stress, HEK293T cells were transfected with the expression constructs and 24 h later, oxidative stress was induced by adding fresh media containing either 250 μM H2O2 or 50 μM tBHQ. The tBHQ was dissolved in ethanol, and control cells were cultured with an equivalent concentration of ethanol (0.1%). The cells were cultured for an additional 4 or 24 h before harvesting and preparing the cell extracts.
SDS-PAGE and western analyses
An aliquot from each of the primary cell suspensions in PBS was supplemented with SDS to a final concentration of 0.5%, and these aliquots were immediately heat denatured at 100°C. The protein concentration of each total cell lysate was determined with the Pierce BCA Protein Assay Kit, and 5 μg of total protein from each transfectant was incubated at 100°C for 10 min in Laemmli sample buffer (2% SDS, 5% glycerol, 5% 2-mercaptoethanol, 0.002% bromphenol blue, 62.5 mM Tris-HCl, pH 6.8) and resolved on 10% SDS-polyacrylamide gels. Resolved proteins were then electroblotted onto nitrocellulose membranes and subjected to western analyses against hGGT1 and hGGT2, as previously described (53). For Nrf2 western analyses, nuclear fractions were prepared with the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific, Waltham, MA), and immunoblots were conducted on 10 μg aliquots of nuclear proteins as indicated earlier, using a previously described rabbit polyclonal antibody against human Nrf2 diluted 1:250 in TBST (56).
Deglycosylation of GGT proteins by N-glycosidase F
Forty microgram aliquots of total protein from each of the SDS-solubilized HEK293T transfectant lysates were diluted to 40 μl with PBS+0.5% 3-[(3-cholamidopropyl)dimethyl-ammonio]-1-propanesulfonate (CHAPS) and supplemented with SDS to 0.5%. The samples were heat denatured at 100°C for 10 min and cooled to room temperature. Each sample was supplemented with protease inhibitors (1 μg/ml aprotinin, 1 μM leupeptin) and 20 units of N-glycosidase F (PNGaseF, EC 3.5.1.52; New England BioLabs, Ipswich, MA) for N-deglycosylation of the glycoproteins as previously described (53). Western blot analyses were then carried out on the deglycosylated samples as described earlier.
RT-PCR analysis
RNA was extracted from P100 plates of transfected HEK293T cells using the Trizol reagent as directed by the manufacturer's protocol (Invitrogen). Specific primers were designed against common sequences in both hGGT1 and each of the three hGGT2 cDNA variants using the Primer3 software (43), and the specificity of the primers was assessed in silico using the BLAST-Like Alignment Tool [BLAT, (26), Supplementary Table S2]. Reverse transcription and PCR amplification were conducted in staged one-step reactions, using the GeneAmp Gold RNA PCR Core kit, according to the manufacturer's protocol (Applied Biosystems, Carlsbad, CA). Amplified cDNAs were visualized in a 1.0% agarose gel stained with ethidium bromide. A 1Kb Plus DNA ladder (Invitrogen) was loaded on each gel for validation of product sizes, and β-actin amplification was conducted in parallel to ensure equivalent loading.
GGT activity assays
The transfected cells were harvested and then lysed in ice cold PBS containing 0.5% CHAPS and protease inhibitors (1 μg/ml aprotinin, 1 μM leupeptin). Insoluble material was pelleted at 15,000 g for 15 min at 4°C. The soluble fractions contained GGT eluted from the membrane. Transpeptidation assays contained 3 mM L-γ glutamylparanitroanalide (L-GpNA; Sigma, St. Louis, MO) as the substrate and 40 mM glycylglycine (glygly; Sigma) as the acceptor and were conducted as previously described (27). For hydrolysis assays D-γ-glutamyl-paranitroanalide (D-GpNA; Bachem, Torrance, CA) was used as the substrate (48). One unit of GGT activity was defined as the amount of enzyme that released 1 μmol of paranitroaniline/min at 37°C, pH 7.4. Unless otherwise stated, reported specific activities refer to those that were measured using the transpeptidation assay. The concentration of protein in the soluble fractions was determined with the Pierce BCA Protein Assay Kit (ThermoScientific, Rockford, IL).
Data analysis
Samples in each experiment were analyzed in triplicate. Each GGT allele was evaluated in two or more independent experiments. Graphs, averages, and standard deviations were calculated using Prism GraphPad Software (San Diego, CA). Data were calculated as the mean±standard deviation of at least three independent determinations.
GGT subcellular localization
Glass coverslips in P35 plates were seeded with 2×105 HEK293 cells and cultured in complete DMEM at 37°C and 5% CO2. After 24 h, each plate of cells was transfected with 6 μg of the appropriate GGT construct as described earlier. Fresh DMEM media were added to each plate after 18 h, and the transfectants were cultured continuously at 37°C and 5% CO2 thereafter. The cells were fixed 48 h after the transfection and then subjected to immunofluorescence, as previously described (20). The mouse monoclonal ER marker, anti-calnexin (ab31290, Abcam, Cambridge, MA; diluted 1:500 in blocking agent), and affinity-purified GGT129 (diluted 1:100 in blocking agent, equivalent to a 1:1300 dilution relative to the serum), a rabbit polyclonal antibody against a 19-amino-acid peptide that is conserved in the C-terminus of the large subunit of both hGGT1 and hGGT2 (14), were used as the primary antibodies. The secondary antibodies were AlexaFluor 488-conjugated goat anti-rabbit IgG and AlexaFluor 568-conjugated goat anti-mouse IgG (each diluted 1:200; both from Invitrogen-Molecular Probes, Carlsbad, CA). Rinsed coverslips containing labeled cells were mounted onto glass slides using Prolong Gold+4′-6-diamidino-2-phenylindole (DAPI; Invitrogen-Molecular Probes). Fluorescent images of labeled cells were captured with an Olympus IX70 epifluorescence microscope (Olympus America, Center Valley, PA) and a QICAM camera controlled by QCapture software (QImaging, Surrey, British Columbia, Canada). Exposure times for GGT images differed among the samples. The reduced levels of GGT2 proteins compared with hGGT1 required much longer exposures to visulalize their subcellular localization. The exposure time for calnexin and DAPI were held constant for all samples. Figures were prepared by calibrating image scale, exporting to Photoshop (Adobe, Mountain View, CA), and adjusting brightness and contrast for each image individually to highlight specific labeling.
Supplementary Material
Abbreviations Used
- ER
endoplasmic reticulum
- GGT1
γ-glutamyltranspeptidase 1
- GGT2
γ-glutamyltranspeptidase 2
- hGGT1
human γ-glutamyltranspeptidase 1
- Ntn
N-terminal nucleophile
- tBHQ
tert-butylhydroquinone
Acknowledgments
The authors would like to thank Dr. Scott Plafker and Kendra Plafker (Oklahoma Medical Research Foundation, Oklahoma City, OK) for their assistance with the oxidative stress experiments and for their kind gift of the rabbit polyclonal human Nrf2 antibody. This work was supported by National Institutes of Health Grants RO1 CA57530 and R56 CA57530 (M.H.H.), OCAST HR11-085 (M.H.H.), and F32 CA128338 (M.B.W.).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Auman JT. Church R. Lee SY. Watson MA. Fleshman JW. McLeod HL. Celecoxib pre-treatment in human colorectal adenocarcinoma patients is associated with gene expression alterations suggestive of diminished cellular proliferation. Eur J Cancer. 2008;44:1754–1760. doi: 10.1016/j.ejca.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baginski L. Tachon G. Falson F. Patton JS. Bakowsky U. Ehrhardt C. Reverse transcription polymerase chain reaction (RT-PCR) analysis of proteolytic enzymes in cultures of human respiratory epithelial cells. J Aerosol Med Pulm Drug Deliv. 2011;24:89–101. doi: 10.1089/jamp.2010.0842. [DOI] [PubMed] [Google Scholar]
- 3.Boanca G. Sand A. Barycki JJ. Uncoupling the enzymatic and autoprocessing activities of Helicobacter pylori gamma-glutamyltranspeptidase. J Biol Chem. 2006;281:19029–19037. doi: 10.1074/jbc.M603381200. [DOI] [PubMed] [Google Scholar]
- 4.Brannigan JA. Dodson G. Duggleby HJ. Moody PC. Smith JL. Tomchick DR. Murzin AG. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 5.Capraro MA. Hughey RP. Use of acivicin in the determination of rate constants for turnover of rat renal gamma-glutamyltranspeptidase. J Biol Chem. 1985;260:3408–3412. [PubMed] [Google Scholar]
- 6.Castonguay R. Halim D. Morin M. Furtos A. Lherbet C. Bonneil E. Thibault P. Keillor JW. Kinetic characterization and identification of the acylation and glycosylation sites of recombinant human gamma-glutamyltranspeptidase. Biochemistry. 2007;46:12253–12262. doi: 10.1021/bi700956c. [DOI] [PubMed] [Google Scholar]
- 7.Chen R. Jiang X. Sun D. Han G. Wang F. Ye M. Wang L. Zou H. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J Proteome Res. 2009;8:651–661. doi: 10.1021/pr8008012. [DOI] [PubMed] [Google Scholar]
- 8.Chevez-Barrios P. Wiseman AL. Rojas E. Ou CN. Lieberman MW. Cataract development in gamma-glutamyl transpeptidase-deficient mice. Exp Eye Res. 2000;71:575–582. doi: 10.1006/exer.2000.0913. [DOI] [PubMed] [Google Scholar]
- 9.Curthoys NP. Hughey RP. Characterization and Physiological-function of rat renal gamma-glutamyl-transpeptidase. Enzyme. 1979;24:383–403. doi: 10.1159/000458694. [DOI] [PubMed] [Google Scholar]
- 10.DeNicola GM. Karreth FA. Humpton TJ. Gopinathan A. Wei C. Frese K. Mangal D. Yu KH. Yeo CJ. Calhoun ES. Scrimieri F. Winter JM. Hruban RH. Iacobuzio-Donahue C. Kern SE. Blair IA. Tuveson DA. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elce JS. Broxmeyer B. Gamma-glutamyltransferase of rat-kidney—simultaneous assay of hydrolysis and transfer-reactions with [glutamate-C-14]glutathione. Biochem J. 1976;153:223–232. doi: 10.1042/bj1530223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emdin M. Pompella A. Paolicchi A. Gamma-glutamyltransferase, atherosclerosis, and cardiovascular disease: triggering oxidative stress within the plaque. Circulation. 2005;112:2078–2080. doi: 10.1161/CIRCULATIONAHA.105.571919. [DOI] [PubMed] [Google Scholar]
- 13.Graham FL. van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 14.Hanigan MH. Frierson HF., Jr. Immunohistochemical detection of gamma-glutamyl transpeptidase in normal human tissue. J Histochem Cytochem. 1996;44:1101–1108. doi: 10.1177/44.10.8813074. [DOI] [PubMed] [Google Scholar]
- 15.Hanigan MH. Frierson HF., Jr. Swanson PE. De Young BR. Altered expression of gamma-glutamyl transpeptidase in human tumors. Hum Pathol. 1999;30:300–305. doi: 10.1016/s0046-8177(99)90009-6. [DOI] [PubMed] [Google Scholar]
- 16.Hanigan MH. Gallagher BC. Townsend DM. Gabarra V. Gamma-glutamyl transpeptidase accelerates tumor growth and increases the resistance of tumors to cisplatin in vivo. Carcinogenesis. 1999;20:553–559. doi: 10.1093/carcin/20.4.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanigan MH. Ricketts WA. Extracellular glutathione is a source of cysteine for cells that express gamma-glutamyl transpeptidase. Biochemistry. 1993;32:6302–6306. doi: 10.1021/bi00075a026. [DOI] [PubMed] [Google Scholar]
- 18.Heisterkamp N. Groffen J. Duplication of the bcr and gamma-glutamyl transpeptidase genes. Nucleic Acids Res. 1988;16:8045–8056. doi: 10.1093/nar/16.16.8045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heisterkamp N. Groffen J. Warburton D. Sneddon TP. The human gamma-glutamyltransferase gene family. Hum Genet. 2008;123:321–332. doi: 10.1007/s00439-008-0487-7. [DOI] [PubMed] [Google Scholar]
- 20.Howard EW. Crider BJ. Updike DL. Bullen EC. Parks EE. Haaksma CJ. Sherry DM. Tomasek JJ. MMP-2 expression by fibroblasts is suppressed by the myofibroblast phenotype. Exp Cell Res. 2012;318:1542–1553. doi: 10.1016/j.yexcr.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu X. Legler PM. Khavrutskii I. Scorpio A. Compton JR. Robertson KL. Friedlander AM. Wallqvist A. Probing the donor and acceptor substrate specificity of the gamma-glutamyl transpeptidase. Biochemistry. 2012;51:1199–1212. doi: 10.1021/bi200987b. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda Y. Fujii J. Anderson ME. Taniguchi N. Meister A. Involvement of Ser-451 and Ser-452 in the catalysis of human gamma-glutamyl transpeptidase. J Biol Chem. 1995;270:22223–22228. doi: 10.1074/jbc.270.38.22223. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda Y. Fujii J. Taniguchi N. Meister A. Human gamma-glutamyl transpeptidase mutants involving conserved aspartate residues and the unique cysteine residue of the light subunit. J Biol Chem. 1995;270:12471–12475. [PubMed] [Google Scholar]
- 24.Ikeda Y. Taniguchi N. Gene expression of gamma-glutamyltranspeptidase. Methods Enzymol. 2005;401:408–425. doi: 10.1016/S0076-6879(05)01025-6. [DOI] [PubMed] [Google Scholar]
- 25.Jean JC. Liu Y. Brown LA. Marc RE. Klings E. Joyce-Brady M. Gamma-glutamyl transferase deficiency results in lung oxidant stress in normoxia. Am J Physiol Lung Cell Mol Physiol. 2002;283:L766–L776. doi: 10.1152/ajplung.00250.2000. [DOI] [PubMed] [Google Scholar]
- 26.Kent WJ. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King JB. West MB. Cook PF. Hanigan MH. A novel, species-specific class of uncompetitive inhibitors of gamma-glutamyl transpeptidase. J Biol Chem. 2009;284:9059–9065. doi: 10.1074/jbc.M809608200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinlough CL. Poland PA. Bruns JB. Hughey RP. Gamma-glutamyltranspeptidase: disulfide bridges, propeptide cleavage, and activation in the endoplasmic reticulum. Methods Enzymol. 2005;401:426–449. doi: 10.1016/S0076-6879(05)01026-8. [DOI] [PubMed] [Google Scholar]
- 29.Lieberman MW. Wiseman AL. Shi ZZ. Carter BZ. Barrios R. Ou CN. Chevez-Barrios P. Wang Y. Habib GM. Goodman JC. Huang SL. Lebovitz RM. Matzuk MM. Growth retardation and cysteine deficiency in gamma-glutamyl transpeptidase-deficient mice. Proc Natl Acad Sci U S A. 1996;93:7923–7926. doi: 10.1073/pnas.93.15.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowry MH. McAllister BP. Jean JC. Brown LA. Hughey RP. Cruikshank WW. Amar S. Lucey EC. Braun K. Johnson P. Wight TN. Joyce-Brady M. Lung lining fluid glutathione attenuates IL-13-induced asthma. Am J Respir Cell Mol Biol. 2008;38:509–516. doi: 10.1165/rcmb.2007-0128OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCracken AA. Brodsky JL. Evolving questions and paradigm shifts in endoplasmic-reticulum-associated degradation (ERAD) Bioessays. 2003;25:868–877. doi: 10.1002/bies.10320. [DOI] [PubMed] [Google Scholar]
- 32.Mehdi K. Penninckx MJ. An important role for glutathione and gamma-glutamyltranspeptidase in the supply of growth requirements during nitrogen starvation of the yeast Saccharomyces cerevisiae. Microbiology. 1997;143Pt 6:1885–1889. doi: 10.1099/00221287-143-6-1885. [DOI] [PubMed] [Google Scholar]
- 33.Miao H. Chen L. Riordan SM. Li W. Juarez S. Crabb AM. Lukas TJ. Du P. Lin SM. Wise A. Agapova OA. Yang P. Gu CC. Hernandez MR. Gene expression and functional studies of the optic nerve head astrocyte transcriptome from normal African Americans and Caucasian Americans donors. PLoS One. 2008;3:e2847. doi: 10.1371/journal.pone.0002847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moon DO. Kim BY. Jang JH. Kim MO. Jayasooriya RG. Kang CH. Choi YH. Moon SK. Kim WJ. Ahn JS. Kim GY. K-RAS transformation in prostate epithelial cell overcomes H(2)O(2)-induced apoptosis via upregulation of gamma-glutamyltransferase-2. Toxicol In Vitro. 2012;26:429–434. doi: 10.1016/j.tiv.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 35.Morrow AL. Williams K. Sand A. Boanca G. Barycki JJ. Characterization of Helicobacter pylori gamma-glutamyltranspeptidase reveals the molecular basis for substrate specificity and a critical role for the tyrosine 433-containing loop in catalysis. Biochemistry. 2007;46:13407–13414. doi: 10.1021/bi701599e. [DOI] [PubMed] [Google Scholar]
- 36.Oinonen C. Rouvinen J. Structural comparison of Ntn-hydrolases. Protein Sci. 2000;9:2329–2337. doi: 10.1110/ps.9.12.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okada T. Suzuki H. Wada K. Kumagai H. Fukuyama K. Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate. Proc Natl Acad Sci U S A. 2006;103:6471–6476. doi: 10.1073/pnas.0511020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osada M. Park HL. Nagakawa Y. Yamashita K. Fomenkov A. Kim MS. Wu G. Nomoto S. Trink B. Sidransky D. Differential recognition of response elements determines target gene specificity for p53 and p63. Mol Cell Biol. 2005;25:6077–6089. doi: 10.1128/MCB.25.14.6077-6089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pandur S. Pankiv S. Johannessen M. Moens U. Huseby NE. Gamma-glutamyltransferase is upregulated after oxidative stress through the Ras signal transduction pathway in rat colon carcinoma cells. Free Radic Res. 2007;41:1376–1384. doi: 10.1080/10715760701739488. [DOI] [PubMed] [Google Scholar]
- 40.Pawlak A. Lahuna O. Bulle F. Suzuki A. Ferry N. Siegrist S. Chikhi N. Chobert MN. Guellaen G. Laperche Y. gamma-Glutamyl transpeptidase: a single copy gene in the rat and a multigene family in the human genome. J Biol Chem. 1988;263:9913–9916. [PubMed] [Google Scholar]
- 41.Pawlak A. Wu SJ. Bulle F. Suzuki A. Chikhi N. Ferry N. Baik JH. Siegrist S. Guellaen G. Different gamma-glutamyl transpeptidase mRNAs are expressed in human liver and kidney. Biochem Biophys Res Commun. 1989;164:912–918. doi: 10.1016/0006-291x(89)91545-3. [DOI] [PubMed] [Google Scholar]
- 42.Rojas E. Valverde M. Kala SV. Kala G. Lieberman MW. Accumulation of DNA damage in the organs of mice deficient in gamma-glutamyltranspeptidase. Mutat Res. 2000;447:305–316. doi: 10.1016/s0027-5107(99)00191-8. [DOI] [PubMed] [Google Scholar]
- 43.Rozen S. Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 44.Sluik D. Beulens JW. Weikert C. van Dieren S. Spijkerman AM. van der AD. Fritsche A. Joost HG. Boeing H. Nothlings U. Gamma-glutamyltransferase, cardiovascular disease and mortality in individuals with diabetes mellitus. Diabetes Metab Res Rev. 2012;28:284–288. doi: 10.1002/dmrr.2261. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki H. Hashimoto W. Kumagai H. Escherichia coli K-12 can utilize an exogenous gamma-glutamyl peptide as an amino acid source, for which gamma-glutamyltranspeptidase is essential. J Bacteriol. 1993;175:6038–6040. doi: 10.1128/jb.175.18.6038-6040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki H. Kumagai H. Autocatalytic processing of gamma-glutamyltranspeptidase. J Biol Chem. 2002;277:43536–43543. doi: 10.1074/jbc.M207680200. [DOI] [PubMed] [Google Scholar]
- 47.Tate SS. Single-chain precursor of renal gamma-glutamyl transpeptidase. FEBS Lett. 1986;194:33–38. doi: 10.1016/0014-5793(86)80046-1. [DOI] [PubMed] [Google Scholar]
- 48.Thompson GA. Meister A. Hydrolysis and transfer reactions catalyzed by gamma-glutamyl transpeptidase; evidence for separate substrate sites and for high affinity of L-cystine. Biochem Biophys Res Commun. 1976;71:32–36. doi: 10.1016/0006-291x(76)90245-x. [DOI] [PubMed] [Google Scholar]
- 49.Thompson JD. Higgins DG. Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wada K. Irie M. Suzuki H. Fukuyama K. Crystal structure of the halotolerant gamma-glutamyltranspeptidase from Bacillus subtilis in complex with glutamate reveals a unique architecture of the solvent-exposed catalytic pocket. FEBS J. 2010;277:1000–1009. doi: 10.1111/j.1742-4658.2009.07543.x. [DOI] [PubMed] [Google Scholar]
- 51.West M. Kang PS. Klouckova I. Feasley C. West C. Novotny M. Mechref Y. Hanigan M. N-Glycosylation of Human Liver and Kidney Gamma-Glutamyl Transpeptidase is Strikingly Different at all Glycosylation Sites. Glycobiology. 2009;19:1332–1332. [Google Scholar]
- 52.West MB. Segu ZM. Feasley CL. Kang P. Klouckova I. Li CL. Novotny MV. West CM. Mechref Y. Hanigan MH. Analysis of site-specific glycosylation of renal and hepatic gamma-glutamyl transpeptidase from normal human tissue. J Biol Chem. 2010;285:29511–29524. doi: 10.1074/jbc.M110.145938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.West MB. Wickham S. Quinalty LM. Pavlovicz RE. Li C. Hanigan MH. Autocatalytic cleavage of human gamma-glutamyl transpeptidase is highly dependent on N-glycosylation at asparagine 95. J Biol Chem. 2011;286:28876–28888. doi: 10.1074/jbc.M111.248823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams DB. Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J Cell Sci. 2006;119:615–623. doi: 10.1242/jcs.02856. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H. Forman HJ. Redox regulation of gamma-glutamyl transpeptidase. Am J Respir Cell Mol Biol. 2009;41:509–515. doi: 10.1165/rcmb.2009-0169TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ziady AG. Sokolow A. Shank S. Corey D. Myers R. Plafker S. Kelley TJ. Interaction with CREB binding protein modulates the activities of Nrf2 and NF-kappaB in cystic fibrosis airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2012;302:L1221–L1231. doi: 10.1152/ajplung.00156.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.